Generating extrema set
Updated 5/27/2020
Written by Reed Stein.
This is a wrapper script that performs everything below including JK's "gen_extrema.py" script. It requires a SMILES file and the number of molecules you would like from each molecular weight/cLogP tranche. Make sure to source python3
source /nfs/soft/python/current/env.csh
Then run:
python ~rstein/zzz.scripts/TLDR_DUDE/0001_db2_map_tranche_collect_db2_gz.py -s {SMILES_FILES}
You can also specify number of molecules to return. The default is 500.
python ~rstein/zzz.scripts/TLDR_DUDE/0001_db2_map_tranche_collect_db2_gz.py -s {SMILES_FILES} -n 1000
This script will find the interquartile range of molecular weight/cLogP properties of your input SMILES ligands. Then it will retrieve db2.gz files for all -2, -1, 0, +1, +2 charges for each MWT/cLogP tranche.
Once this finishes running, as below:
cat *_charge_tranches.list > extrema_set.list
which is the split_database_index you would then use for docking.
Written by Jiankun Lyu, 2019/10/12
The main purpose of the extrema set is to test the charge preference of your docking setup and to make sure that you don't over-optimize your docking setup with property-matched(charged-matched) decoys generated by DUDE. This is also a sanity check of your docking setup, put it on your checklist!
extrema_set_gen------- working
|
|------ ZINC-downloader-3D-minu2.database_index
|
|------ ZINC-downloader-3D-minu1.database_index
|
|------ ZINC-downloader-3D-neutral.database_index
|
|------ ZINC-downloader-3D-plus1.database_index
|
|------ ZINC-downloader-3D-plus2.database_index
1) Make those directories above.
mkdir extrema_set_gen cd extrema_set_gen mkdir working
2) Download databases index from ZINC with different charge types
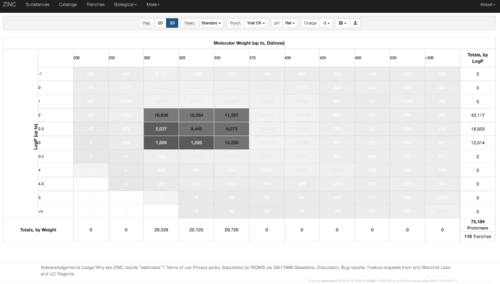
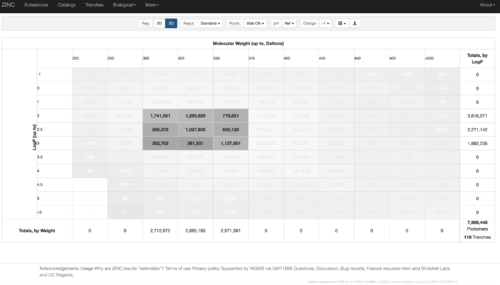
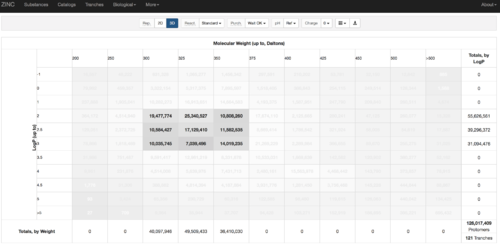
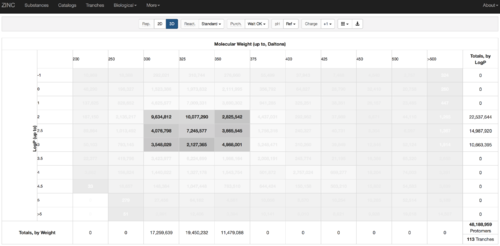
2.1) Go to ZINC http://zinc15.docking.org/tranches/home/#
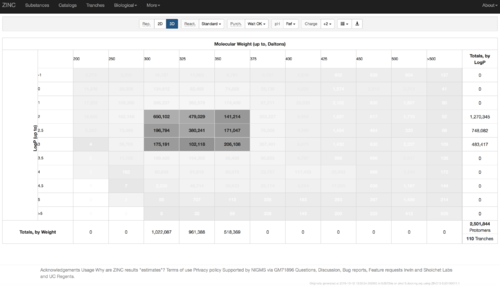
2.2) Choose the tranches you want to generate extrema set for testing the charge preference. The goldilocks set has been chosen here as an example.
2.3) download the databases index file for each charge type
2.4) download the files above and save it as ZINC-downloader-3D-(charge-type).database_index, then upload the file to the working directory. In the working directory, you are supposed to have 5 files with names: ZINC-downloader-3D-minu2.database_index, ZINC-downloader-3D-minu1.database_index, ZINC-downloader-3D-neutral.database_index, ZINC-downloader-3D-plus1.database_index and ZINC-downloader-3D-plus2.database_index.
3) Run extrema set generation on 5 different charge types
python /mnt/nfs/ex5/work/jklyu/sigma2/gen_extrema/script/gen_extrema.py First input: the DB index from ZINC15 Second input: the prefix of the ligand charge Third input: the lower bound of number of molecules for each tranche
python /mnt/nfs/ex5/work/jklyu/sigma2/gen_extrema/script/gen_extrema.py ZINC-downloader-3D-plus2.database_index 'plus2' 100 > log_plus2 & python /mnt/nfs/ex5/work/jklyu/sigma2/gen_extrema/script/gen_extrema.py ZINC-downloader-3D-plus1.database_index 'plus1' 100 > log_plus1 & python /mnt/nfs/ex5/work/jklyu/sigma2/gen_extrema/script/gen_extrema.py ZINC-downloader-3D-neutral.database_index '0' 100 > log_0 & python /mnt/nfs/ex5/work/jklyu/sigma2/gen_extrema/script/gen_extrema.py ZINC-downloader-3D-minus1.database_index 'minus1' 100 > log_minus1 & python /mnt/nfs/ex5/work/jklyu/sigma2/gen_extrema/script/gen_extrema.py ZINC-downloader-3D-minus2.database_index 'minus2' 100 > log_minus2 &
4) Output files
4.1) (charge-type)_tranche_summary.txt. The file contains how many molecules has been selected from each tranche. The section below is an example:
EF 430 DF 337 ED 293 DD 181 DE 120 CF 112 CE 131 CD 272 EE 118 1994
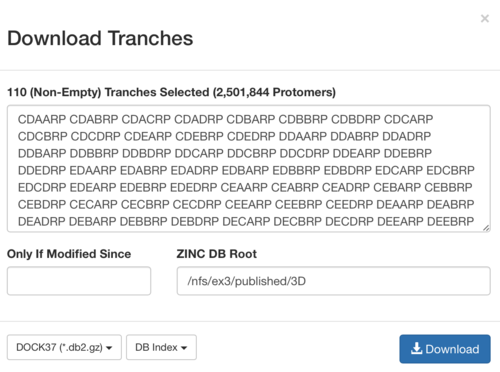
4.2) (charge-type)_charge_tranches.list. The file contains all the db2 indexes that have been selected from the extrema generation.
5) Combine all the db2 indexes generated by the script.
cat *_charge_tranches.list > extrema_set.list
6) Use the extrema_set.list as .sdi to set up your docking screen then run it.