Category:SEA: Difference between revisions
| (6 intermediate revisions by the same user not shown) | |||
| Line 21: | Line 21: | ||
=== Compounds vs Targets === | === Compounds vs Targets === | ||
==== Compound vs Library screen ==== | ==== Compound vs Library screen ==== | ||
[[File:compound_vs_library.png| | [[File:compound_vs_library.png|700px|right|alt text]] | ||
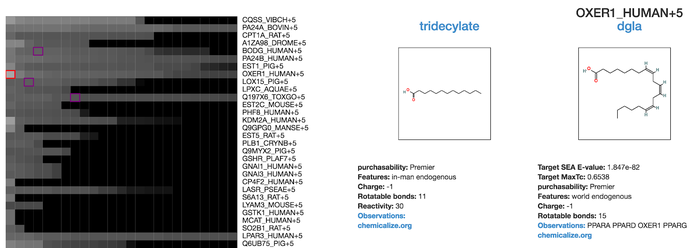
* User has one or more compounds of interest, such as one that has come out of a phenotypic screen and wants to predict its mechanism of action. | * User has one or more compounds of interest, such as one that has come out of a phenotypic screen and wants to predict its mechanism of action. | ||
* The user inputs the smiles and optionally compound IDs and selects a library (e.g. ChEMBL27 by protein) (<b>/search</b> route) | * The user inputs the smiles and optionally compound IDs and selects a library (e.g. ChEMBL27 by protein) (<b>/search</b> route) | ||
| Line 30: | Line 30: | ||
** Similar targets to predicted target (feature request) | ** Similar targets to predicted target (feature request) | ||
** Sequence similarity and ligand similarity trees for predicted targets (feature request) | ** Sequence similarity and ligand similarity trees for predicted targets (feature request) | ||
==== Compounds vs Library screen ==== | ==== Compounds vs Library screen ==== | ||
[[File:compound_vs_target.png|300px|right|alt text]] | |||
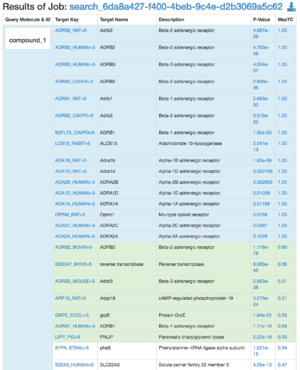
* Same as above except instead of going directly to <b>/compound_vs_target route</b>, go to a summary page for each compound (<b>/results</b> route) | * Same as above except instead of going directly to <b>/compound_vs_target route</b>, go to a summary page for each compound (<b>/results</b> route) | ||
==== Compound(s) vs Specified Library targets ==== | ==== Compound(s) vs Specified Library targets ==== | ||
| Line 39: | Line 41: | ||
=== Target vs Targets === | === Target vs Targets === | ||
==== Target vs Target ==== | ==== Target vs Target ==== | ||
[[File:target_vs_target.png|700px|right|alt text]] | |||
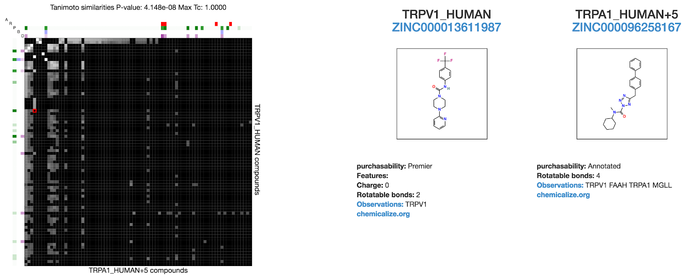
* The user wants to compare the ligand similarity between two targets | * The user wants to compare the ligand similarity between two targets | ||
* The user inputs the target id one as the reference target and one as the query target(<b>/search</b> route) | * The user inputs the target id one as the reference target and one as the query target(<b>/search</b> route) | ||
Latest revision as of 23:03, 25 July 2022
| Paragigm | Docking |
|---|---|
| Developer | Shoichet Lab |
| Stable Version | SEA |
| Programming Language | TBD |
| Dependencies | TBD |
SEA, the Similarity Ensemble Approach. The website is: sea.bkslab.org
About
SEA relates proteins based on the set-wise chemical similarity among their ligands. It can be used to rapidly search large compound databases and to build cross-target similarity maps. It is provided by the Shoichet Laboratory in the Department of Pharmaceutical Chemistry at the University of California, San Francisco (UCSF).
Citing SEA
To cite SEA, please reference Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotech 25 (2), 197-206 (2007)[1].
Publications
Use cases
Compounds vs Targets
Compound vs Library screen
- User has one or more compounds of interest, such as one that has come out of a phenotypic screen and wants to predict its mechanism of action.
- The user inputs the smiles and optionally compound IDs and selects a library (e.g. ChEMBL27 by protein) (/search route)
- Returns the page where of predicted targets and the most similar annotated to the query target when a compound in a predicted target is selected, the query target and the reference target are shown on the side(/compound_vs_target route)
- User can evaluate predictions and link
- Additional information about predicted targets (Uniprot or Zinc15 orthologs/genes page)
- Additional information about compounds annotated to predicted targets (Zinc15 substance page)
- Similar targets to predicted target (feature request)
- Sequence similarity and ligand similarity trees for predicted targets (feature request)
Compounds vs Library screen
- Same as above except instead of going directly to /compound_vs_target route, go to a summary page for each compound (/results route)
Compound(s) vs Specified Library targets
- Same as above except instead of returning results for all targets in the library, just return results for the requested targets
Compound(s) vs Custom Targets
- Same as above instead of targets defined in a library, the user specifies a list of target-compound activities that define one or more targets
Target vs Targets
Target vs Target
- The user wants to compare the ligand similarity between two targets
- The user inputs the target id one as the reference target and one as the query target(/search route)
- This returns a ligand similarity matrix where the rows and columns are compounds in each target and when a cell is selected information about the compounds is shown on the side (/target_vs_target route)
Target vs Library screen
- The user wants to find all targets with ligand similarity to a given target in the library
- The user selects the reference library and the query target id from a library
- This returns a results page of for each target in the reference library similar to the query target (/results route)
- Clicking on one of the results take the user to the (/target_vs_target route) described above
Targets vs Library screen
- Same as as above except more than one target are queried and all the results are shown in the results page
Target(s) vs Specific Library Targets
- Same as above but instead of screening against the full reference library a subset of target ids are specified, and results for only those targets are shown.
Custom target(s) vs. [target, library screen, specific library targets]
- Same as above except the user can define the targets by specifying the target name and the active compounds (one pair per line)
Obtaining a private copy of SEA
Please contact support(at)sea*bkslab*org
Other topics
Notes
Caveat Emptor! SEA is provided free-of-charge in the hope that it will be useful, but you must use it at your own risk. We make no guarantees about data confidentiality on this public service website. If you would like to use SEA securely, please contact support+sea*bkslab*org.
To determine whether SEA's predictions for your compounds are already known, we recommend you visit the extensive Ki Database at the NIMH Psychoactive Drug Screening Program[2].
References
Pages in category "SEA"
The following 9 pages are in this category, out of 9 total.