Bootstrap AUC: Difference between revisions
No edit summary |
No edit summary |
||
| Line 8: | Line 8: | ||
Get help information: | Get help information: | ||
python /nfs/home/yingyang/scripts/bootstrap_AUC_violinoplot.py -h | python /nfs/home/yingyang/scripts/bootstrap_AUC_violinoplot.py -h | ||
usage: bootstrap_AUC_violinoplot.py [-h] [-l LIG_FILE] [-d DEC_FILE] [-s1 REF] | usage: bootstrap_AUC_violinoplot.py [-h] [-l LIG_FILE] [-d DEC_FILE] [-s1 REF] | ||
[-s2 NEW [NEW ...]] [-sys SYS_NAME] | [-s2 NEW [NEW ...]] [-sys SYS_NAME] | ||
[-n {10..5000}] [-m {AUC,logAUC,both}] | [-n {10..5000}] [-m {AUC,logAUC,both}] | ||
[-t {ttest_ind,ttest_rel,ztest}] | [-t {ttest_ind,ttest_rel,ztest}] | ||
optional arguments: | optional arguments: | ||
-h, --help show this help message and exit | -h, --help show this help message and exit | ||
| Line 38: | Line 36: | ||
python ~/scripts/bootstrap_AUC_violinoplot.py -s1 D4_dock/ -s2 amber/ rescore/ freeform/ -n 100 \ | python ~/scripts/bootstrap_AUC_violinoplot.py -s1 D4_dock/ -s2 amber/ rescore/ freeform/ -n 100 \ | ||
-l ligands.name -d decoys.name | -l ligands.name -d decoys.name | ||
-t ztest | -m both -t ztest | ||
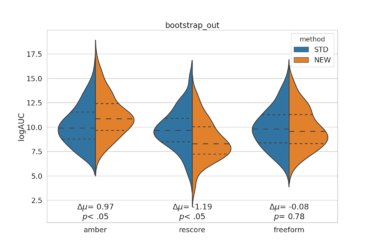
[[File:bootstrap_out_logAUC.png|thumb|center|375px]] | |||
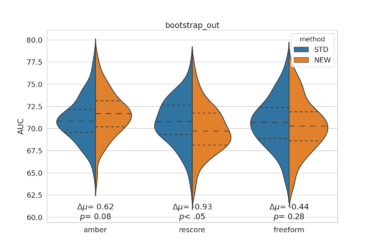
[[File:bootstrap_out_AUC.png|thumb|center|375px]] | |||
Revision as of 01:13, 26 March 2020
3/25/2020 Ying Yang
New violinplot and more statistical tests (Paired T-test, Unpaired T-test, Z-test)
First, set environment variable
source /nfs/home/yingyang/.cshrc_opencadd
Get help information:
python /nfs/home/yingyang/scripts/bootstrap_AUC_violinoplot.py -h
usage: bootstrap_AUC_violinoplot.py [-h] [-l LIG_FILE] [-d DEC_FILE] [-s1 REF]
[-s2 NEW [NEW ...]] [-sys SYS_NAME]
[-n {10..5000}] [-m {AUC,logAUC,both}]
[-t {ttest_ind,ttest_rel,ztest}]
optional arguments:
-h, --help show this help message and exit
-l LIG_FILE, -lig LIG_FILE
File contain ligand names. (default: ligands.name)
-d DEC_FILE, -dec DEC_FILE
File contain decoys names. (default: decoys.name)
-s1 REF Folder with method 1 (ref) score file:
extract_all.sort.uniq.txt (default: None)
-s2 NEW [NEW ...] Folder(s) with new method(s) score.
extract_all.sort.uniq.txt (default: None)
-sys SYS_NAME Name of system to add on plot (default: None)
-n {10..5000}, -num {10..5000}
Number of bootstrap replicate. (default: 50)
-m {AUC,logAUC,both}, -metrics {AUC,logAUC,both}
choose to use AUC or logAUC as the metrics. (default:
both)
-t {ttest_ind,ttest_rel,ztest}, -test {ttest_ind,ttest_rel,ztest}
choose stats test to report p value. Default=
(default: ztest)
Usage example: Compare the AUC and logAUC for three new methods comparing to standard dock. Bootstrap 100 times, and run a Z-test to get p-value.
python ~/scripts/bootstrap_AUC_violinoplot.py -s1 D4_dock/ -s2 amber/ rescore/ freeform/ -n 100 \ -l ligands.name -d decoys.name -m both -t ztest
=========================================================
To test whether the difference in AUC/logAUC between two methods is statistically significant or not, AUC/logAUC of the new developed method(s) against the reference method can be compared with bootstrap.
Files needed:
- ligands.name --> file with ligand names to perform enrichment
- decoys.name --> file with decoy names to perform enrichment
- score file(s) --> extract_all.sort.uniq.txt
First, the anaconda python environment needs to be set:
source /nfs/home/yingyang/.cshrc_anaconda
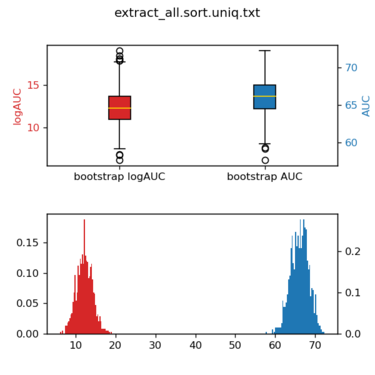
Plot the variation of AUC/logAUC
python /nfs/home/yingyang/work/scripts/bootstrap_AUC.py \ -l ./ligands.name -d ./decoys.name \ -s1 extract_all.sort.uniq.txt \ -p single
The figure will looks like this:
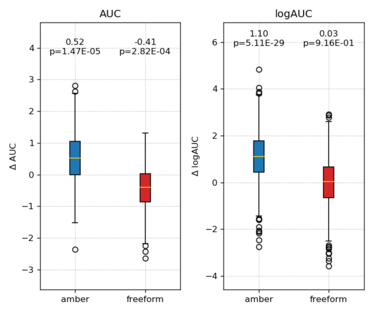
Plot the change(s) in AUC/logAUC against reference score
python /nfs/home/yingyang/work/scripts/bootstrap_AUC.py \ -l ../ligands.name -d ../decoys.name \ -s1 score.standard -s2 score.amber score.freeform \ -p compare
Delta AUC and delta logAUC will be computed and displayed. The p-value from paired t-test indicate if change is statistically significant or not.