Interactive ligands visualizer: Difference between revisions
No edit summary |
No edit summary |
||
| Line 38: | Line 38: | ||
chembl_csv = "mor_chembl_emax.csv" | chembl_csv = "mor_chembl_emax.csv" | ||
activity_name = " | activity_name = "Emax" # The text name of the activity (in this case, Emax) | ||
preprocess_chembl(chembl_csv, activity_name, max_hac=35, max_mw=600, img_folder="mol_images") | preprocess_chembl(chembl_csv, activity_name, max_hac=35, max_mw=600, img_folder="mol_images") | ||
As you can see, you can specify the maximum number of heavy atoms (max_hac) and maximum molecular weight (max_mw) for the ligands to keep. | |||
This will generate two files: a .smi file with the Smiles for all the kept ligands, and .df file which keeps the activity value (Emax here) in dataframe format. | |||
'''Step 4: compute the fingerprints on Gimel''' | |||
Copy the .smi file to gimel, source the DOCK3.7 base, and then run this command (on gimel, not gimel2 or others): | |||
python ~jklyu/zzz.github/ChemInfTools/utils/teb_chemaxon_cheminf_tools/generate_chemaxon_fingerprints.py mor_chembl_emax.smi mor_chembl_emax | |||
This will generate a .fp file, in the present case mor_chembl_emax.fp | |||
'''Step 5: tSNE and interactive visualization''' | |||
Almost done! Copy the .fp file back to your machine, then run part 2 of the example script: | |||
from chemspace_vis.preprocess import make_tsne_from_fingerprints | |||
from chemspace_vis.visualizer import make_visualizer_script | |||
fingerprints_file = "mor_chembl_emax.fp" | |||
make_tsne_from_fingerprints(fingerprints_file) | |||
make_visualizer_script("tsne_data.df", "mol_images", | |||
activity_filename="mor_chembl_emax_activity.df", use_log10=False) | |||
Revision as of 22:45, 20 January 2023
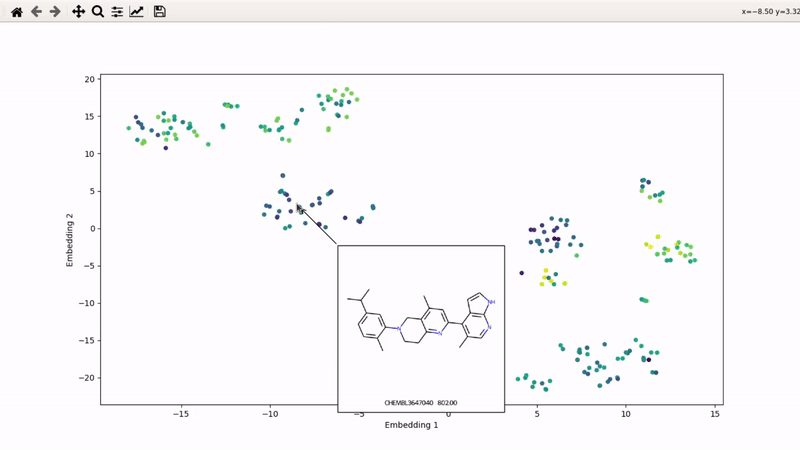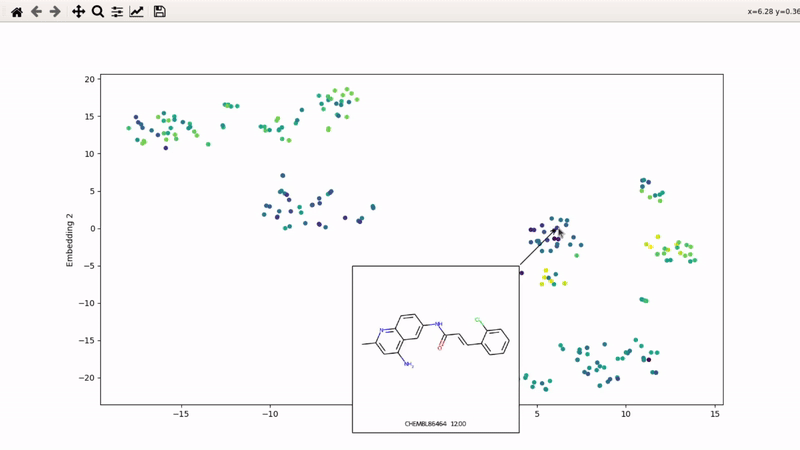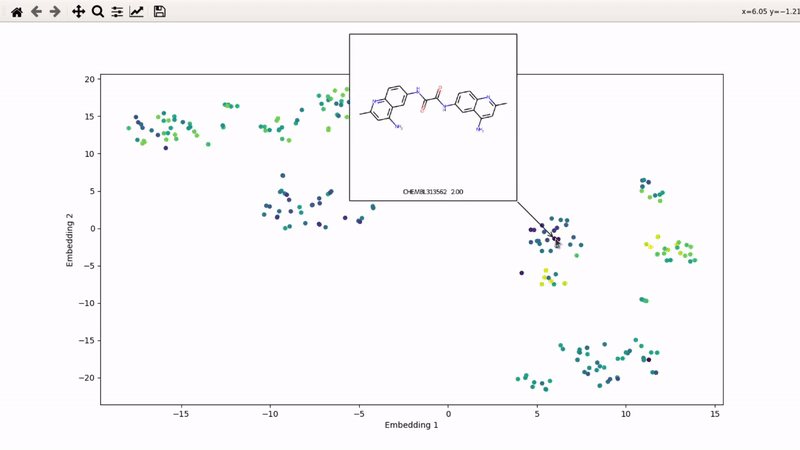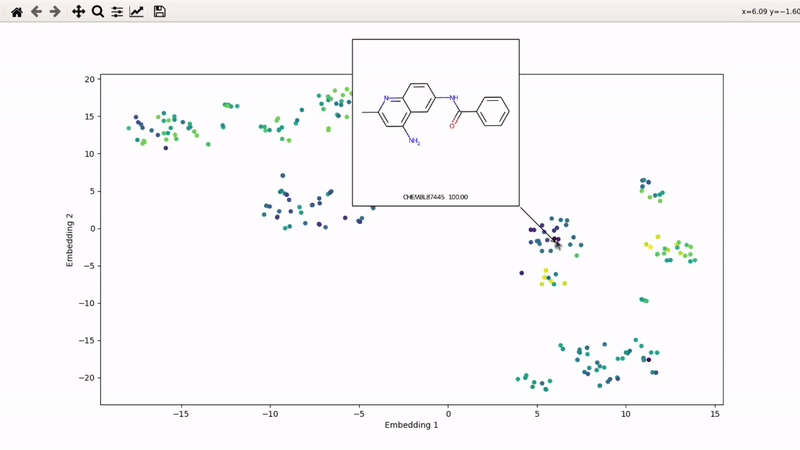
I (Olivier) put together this interactive visualizer to make sure that I don't miss out some chemotypes when coming up with actives at the start of a retrospective campaign. Starting from a downloaded ChEMBL CSV file for a list of ligands, images of each molecule are generated with RDKit and a text file with filtered Smiles is generated. You then need to compute the ECFP fingerprints on Gimel from that file (see below), and then a generated script will show an interactive visualization of the chemical space spanned by the ligands (tSNE), with each molecule shown on mouse hovering.
Step 1: install chemspace_vis package
Make sure you are using Python 3, and then simply:
pip install chemspace_vis
N.B. This only works on Mac and Linux, sorry Windows users (if you exist).
Step 2: obtain ChEMBL CSV file (or use provided example)
Any ChEMBL CSV from a given activity of a given target will do.
You can also clone the example repository, which contains the CSV for mu-opioid ligands with measured Emax and an example script:
git clone https://github.com/gregorpatof/chemspace_vis_example
Just to make things too clear, here is how I obtained that CSV:
Step 3: extract Smiles and activity for given HAC and MW filters
This is accomplished by the preprocess_part1() method in the example script, which runs a single command:
from chemspace_vis.preprocess import preprocess_chembl chembl_csv = "mor_chembl_emax.csv" activity_name = "Emax" # The text name of the activity (in this case, Emax) preprocess_chembl(chembl_csv, activity_name, max_hac=35, max_mw=600, img_folder="mol_images")
As you can see, you can specify the maximum number of heavy atoms (max_hac) and maximum molecular weight (max_mw) for the ligands to keep.
This will generate two files: a .smi file with the Smiles for all the kept ligands, and .df file which keeps the activity value (Emax here) in dataframe format.
Step 4: compute the fingerprints on Gimel
Copy the .smi file to gimel, source the DOCK3.7 base, and then run this command (on gimel, not gimel2 or others):
python ~jklyu/zzz.github/ChemInfTools/utils/teb_chemaxon_cheminf_tools/generate_chemaxon_fingerprints.py mor_chembl_emax.smi mor_chembl_emax
This will generate a .fp file, in the present case mor_chembl_emax.fp
Step 5: tSNE and interactive visualization
Almost done! Copy the .fp file back to your machine, then run part 2 of the example script:
from chemspace_vis.preprocess import make_tsne_from_fingerprints
from chemspace_vis.visualizer import make_visualizer_script
fingerprints_file = "mor_chembl_emax.fp"
make_tsne_from_fingerprints(fingerprints_file)
make_visualizer_script("tsne_data.df", "mol_images",
activity_filename="mor_chembl_emax_activity.df", use_log10=False)