Substructure searching: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
| | | | ||
| | | | ||
------- | ------- scripts ------ submit.csh | ||
| | | | ||
|------ submit_sub_search.csh | |------ submit_sub_search.csh | ||
| Line 25: | Line 25: | ||
cd substructure_searching | cd substructure_searching | ||
mkdir working | mkdir working | ||
mkdir | mkdir scripts | ||
2) Download databases index from ZINC | 2) Download databases index from ZINC | ||
| Line 36: | Line 36: | ||
2.3) download the databases index file | 2.3) download the databases index file | ||
[[File:subsearching_fig2.png|thumb|center|500px|download the databases index file]] | [[File:subsearching_fig2.png|thumb|center|500px|download the databases index file]] | ||
2.4) download the file above and save it as ZINC-downloader-2D-smi.database_index, then upload the file to the working directory | |||
3) Copy scripts from my path. | 3) Copy scripts from my path. | ||
cd | cd scripts | ||
cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/setup_substructure_searching_files.py . | cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/setup_substructure_searching_files.py . | ||
cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/submit.csh . | cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/submit.csh . | ||
| Line 44: | Line 46: | ||
cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/submit_sub_search.csh . | cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/submit_sub_search.csh . | ||
cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/search_multi_substructures.py . | cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/search_multi_substructures.py . | ||
cd ../ | |||
4) Put SMARTS patterns you want to search in the sub_pattern.smarts file and give each SMARTS pattern a unique number or name | |||
Here is an example in the sub_pattern.smarts file | |||
NS(=O)(=O)c1cccc([F,Cl,Br,I])c1[OD1] 1 | |||
NS(=O)(=O)c1cc([F,Cl,Br,I])ccc1[OD1] 2 | |||
5) split the ZINC-downloader-2D-smi.database_index file into chunks | |||
cd working | |||
python ../scripts | |||
Revision as of 18:21, 13 September 2017
Written by Jiankun Lyu, 2017/09/13
The hierarchy of the directories:
substructure_searching----- working
| |
| |------ ZINC-downloader-2D-smi.database_index
| |
| |------ sub_pattern.smarts
|
|
|
------- scripts ------ submit.csh
|
|------ submit_sub_search.csh
|
|------ run_sub_search.csh
|
|------ search_multi_substructures.py
|
|------ setup_substructure_searching_files.py
1) Make those directories above.
mkdir substructure_searching cd substructure_searching mkdir working mkdir scripts
2) Download databases index from ZINC
2.1) Go to ZINC http://zinc15.docking.org/tranches/home/#
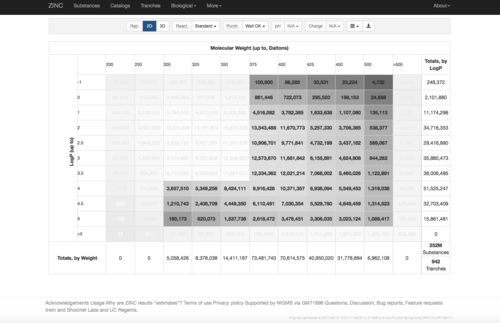
2.2) Choose the tranches you want to do substructure searching
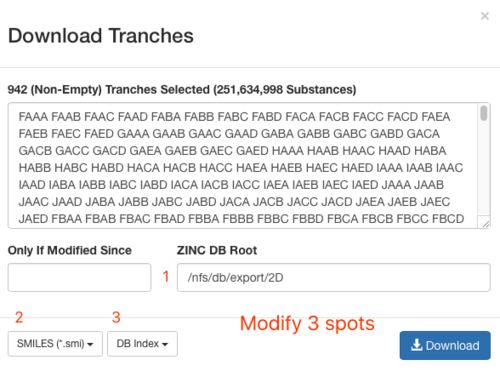
2.3) download the databases index file
2.4) download the file above and save it as ZINC-downloader-2D-smi.database_index, then upload the file to the working directory
3) Copy scripts from my path.
cd scripts cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/setup_substructure_searching_files.py . cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/submit.csh . cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/run_sub_search.csh . cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/submit_sub_search.csh . cp /mnt/nfs/home/jklyu/zzz.script/analogs_searching/multi_sub_searching/search_multi_substructures.py . cd ../
4) Put SMARTS patterns you want to search in the sub_pattern.smarts file and give each SMARTS pattern a unique number or name
Here is an example in the sub_pattern.smarts file NS(=O)(=O)c1cccc([F,Cl,Br,I])c1[OD1] 1 NS(=O)(=O)c1cc([F,Cl,Br,I])ccc1[OD1] 2
5) split the ZINC-downloader-2D-smi.database_index file into chunks
cd working python ../scripts