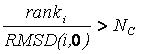Pruning the conformation tree: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
Each candidate is then evaluated for removal based on its rank and RMSD using the inequality shown in Equation 2. If the factor is greater than num_anchors_orients_for_growth or number_confs_for_next_growth, as appropriate, the candidate is removed. | Each candidate is then evaluated for removal based on its rank and RMSD using the inequality shown in Equation 2. If the factor is greater than num_anchors_orients_for_growth or number_confs_for_next_growth, as appropriate, the candidate is removed. | ||
[[Image:Pruning.jpg|center]] | |||
Based on this factor, a configuration with rank 2 and 0.2 Angstroms RMSD is comparable to a configuration with rank 20 and 2.0 Angstroms RMSD. The next best scoring configuration which survives the first pass of removal is then set aside and used as a reference configuration for the second round of pruning, and so on. | Based on this factor, a configuration with rank 2 and 0.2 Angstroms RMSD is comparable to a configuration with rank 20 and 2.0 Angstroms RMSD. The next best scoring configuration which survives the first pass of removal is then set aside and used as a reference configuration for the second round of pruning, and so on. | ||
This pruning method attempts to balance the twin goals of recovering the best scoring and the most different binding configurations without introducing additional user parameters. The pruning method biases its search time towards molecules which sample a more diverse set of binding modes. As the value of num_anchors_orients_for_growth and number_confs_for_next_growth is increased, the anchor-first method approaches an exhaustive search. | This pruning method attempts to balance the twin goals of recovering the best scoring and the most different binding configurations without introducing additional user parameters. The pruning method biases its search time towards molecules which sample a more diverse set of binding modes. As the value of num_anchors_orients_for_growth and number_confs_for_next_growth is increased, the anchor-first method approaches an exhaustive search. | ||
Revision as of 22:43, 22 September 2006
During each cycle of the conformation search, the expanded set of partial configurations is pruned based on the setting of num_anchor_orients_for_growth for anchor orientation pruning and number_confs_for_next_growth for flexibly grown conformations. The pruning attempts to retain the best, most diverse configurations using a top-first pruning method, which proceeds as follows. The configurations are ranked according to score. The top-ranked configuration is set aside and used as a reference configuration for the first round of pruning. All remaining configurations are considered candidates for removal. A root-mean-squared distance (RMSD) between each candidate and the reference configuration is computed.
Each candidate is then evaluated for removal based on its rank and RMSD using the inequality shown in Equation 2. If the factor is greater than num_anchors_orients_for_growth or number_confs_for_next_growth, as appropriate, the candidate is removed.
Based on this factor, a configuration with rank 2 and 0.2 Angstroms RMSD is comparable to a configuration with rank 20 and 2.0 Angstroms RMSD. The next best scoring configuration which survives the first pass of removal is then set aside and used as a reference configuration for the second round of pruning, and so on.
This pruning method attempts to balance the twin goals of recovering the best scoring and the most different binding configurations without introducing additional user parameters. The pruning method biases its search time towards molecules which sample a more diverse set of binding modes. As the value of num_anchors_orients_for_growth and number_confs_for_next_growth is increased, the anchor-first method approaches an exhaustive search.