Omega.parm
Changing settings for OMEGA (OpenEye)
You may be interested to change the default settings for OMEGA (defined in omega.py) when generating conformations for docking. Mabye you want to increase the number of conformations sampled per ligand, or the force field used.
The best way to change the settings for OMEGA is to copy a file called omega.parm into the directory where you want to generate ligand conformations using dbgen.csh. You should find a template in $DOCK_BASE/data/omega.parm
1) Copy omega.parm in your working directory:
cp $DOCK_BASE/data/omega.parm .
(=> if your ligand has no rings, copy and edit omega_noring.parm ($DOCK_BASE/data/omega_noring.parm)
2) Edit and save omega.parm (using the same name) in your favorite editor.
3) Run dbgen.csh with altered settings in omega.parm (has to be in directory where you run dbgen.csh):
dbgen.csh my_ligs.smi
Increase ligand conformation sampling or change the type of force field
An example of omega.parm is shown at the bottom of this page. Settings you may want to consider changing are:
omega.SetEnergyWindow(12.5)
omega.SetMaxConfs(600)
omega.SetRMSThreshold(0.80)
omega.SetBuildForceField('mmff94s_NoEstat')
omega.SetSearchForceField('mmff94s_NoEstat')
- you could increase the energy window: e.g. omega.SetEnergyWindow(25.0)
=> There is no benchmarking of this setting that I am aware of...
- you could increase the number of output conformations: e.g. omega.SetMaxConfs(10000)
=> Molecules in ZINC have a cutoff at 600 conformations, which is certainly on the low side for flexible ligands
- you could lower the RMS threshold between the confs: e.g. omega.SetRMSThreshold(0.40)
=> This results in finer sampling of conformational space
- you could change the force field to included electrostatics: e.g. omega.SetBuildForceField('mmff94s') and omega.SetSearchForceField('mmff94s')
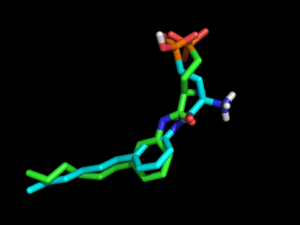
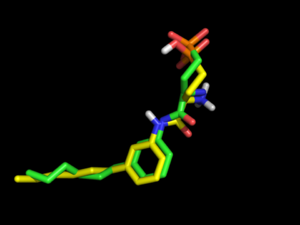
=> Internal benchmarkings on DUDE (by RGC) have shown that electrostatics improves enrichment for many targets. And for ligands exhibiting internal salt bridges, it may be essential to include electrostatics during conformation generation, as shown below in the example of re-docking the S1P1R ligand.
Example of omega.parm
# Example omega toolkit configuration file
# File Parameters
SetCommentEnergy(False)
SetIncludeInput(False)
SetRotorOffset(True)
SetSDEnergy(False)
SetWarts(True)
# 3D Construction Parameters
SetBuildForceField('mmff94s_NoEstat')
SetCanonOrder(True)
SetFixDeleteH(True)
SetDielectric(1.0)
SetExponent(1.0)
SetFixRMS(0.15)
SetFromCT(False)
SetFixMaxMatch(1)
SetFixUniqueMatch(True)
# Structure Enumeration Parameters
SetEnumNitrogen(False)
SetEnumRing(False)
# Torsion Driving Parameters
SetEnergyWindow(12.5)
SetMaxConfs(600)
SetMaxRotors(-1)
SetMaxSearchTime(120.0)
SetRangeIncrement(5)
SetRMSThreshold(0.80)
SetSearchForceField('mmff94s_NoEstat')
SetTorsionDrive(True)
Example of omega_noring.parm
# Omega file with special no ring conformation parameters SetMaxConfs(30)
Acknowledgments
Thanks to Michael M. and RGC.