|
|
| (9 intermediate revisions by 3 users not shown) |
| Line 1: |
Line 1: |
| '''DOCK Blaster Tutorial 1. Dock to Mineralocorticoid receptor''' A[[DOCK Blaster:Tutorials | DOCK Blaster Tutorial]]. | | '''DOCK Blaster Tutorial 1. Dock to Human Thyroid Hormone Receptor Beta-1 (TR beta)''' |
|
| |
|
| [[Image:2aa2.jpg|thumb|right|Mineralocorticoid receptor, PDB 2aa2]] | | A[[DOCK Blaster:Tutorials | DOCK Blaster Tutorial]]. |
|
| |
|
| = Specific Aims =
| |
| This tutorial will show you how to retrospectively dock a crystallographically observed ligand, aldosterone, into the human mineralocorticoid receptor, PDB code
| |
| [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=2AA2 2aa2]. You will also dock a list of annotated actives from the literature, as well as property-matched decoys. Finally, you will prospectively dock the ZINC "fragment-like" library into MR, and pick compounds to test as possible binders. The questions are:
| |
|
| |
|
| * 1. Can DOCK Blaster re-dock the native ligand close to its crystallographically observed position, with a competitive score?
| | ==Step 1== |
| * 2. Can DOCK Blaster enrich known actives from a database of property-matched decoys?
| | a. Browse to the DOCK Blaster main page: |
| * 3. Can DOCK Blaster suggest novel, commercially available ligands for MR?
| | http://blaster.docking.org/ |
| | and select '''Start with a PDB code''' from the '''DOCK''' pull down menu: |
| | http://blaster.docking.org/parser.shtml |
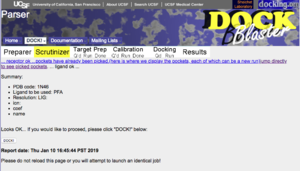
| | b. In the PDB code field, enter 1N46, the PDB code for TR beta. |
| | http://www.ebi.ac.uk/msd-srv/oca/oca-bin/ocashort?id=1N46 |
| | Click on '''DOCK''' to start. |
|
| |
|
| [[Image:Aldosterone.png|thumb|right|Aldostereone, the crystallographically observed ligand of MR in PDB 2aa2]]
| |
|
| |
|
| {{TOCright}}
| | If you are really impatient you can do all of the above |
| | by simply clicking here: |
| | http://blaster.docking.org/cgi-bin/parser.pl?code=1N46 |
|
| |
|
| = Background and Significance = | | ==Step 2== |
| Mineralocorticoid receptor (MR) is a receptor with high affinity for mineralocorticoids. It belongs to the steroid hormone receptor family where the ligand diffuses into cells, interacts with the receptor and results in a single transduction affecting specific gene expression in the nucleus. [http://en.wikipedia.org/wiki/Mineralocorticoid_receptor see Wikipedia].
| | You will get a confirmation page showing that the target has been looked up correctly at the PDB. To proceed, click on "DOCK". Write down the DOCK Blaster Job ID number. You will need it. The docking process is now queued, and may have started if the system is not overloaded. |
| | [[File:DOCK_Blaster_step1a.png|300px|center]] |
|
| |
|
| Hippocampal mineralocorticoid receptors play a major role in the control of the hypothalamus-pituitary-adrena (HPA) axis, and thus MR is a target for drug discovery.
| | ==Step 3== |
| | Click on the link to browse progress at the bottom of the page |
| | http://blaster.docking.org/cgi-bin/jobwatch.pl?job_id=35605&pin= |
| | (replace the job number with your actual job number. This one will not work) |
|
| |
|
|
| |
|
| |
|
| = Preliminary Results =
| | Step 4. When the job has finished, you will be in the "Calibration Done" state as shown in the progress bar with the yellow paint near the top of the page. You will see a report that reports how well docking worked. |
| We will attempt to use DOCK Blaster to answer these questions, using data that has been prepared in advance. If you wish to use DOCK Blaster on your own project, you must prepare data to conform with [[DOCK Blaster:Input Preparation | DOCK Blaster's]] basic requirements. To do this tutorial, please carefully follow the steps below. This tutorial usually takes about two hours, but most of the time is simply waiting for the computers to do the work on our servers.
| |
|
| |
|
| == Protocol 1 - aquire target structure and prepare input files ==
| | Step 5. Interpreting the calibration summary table |
|
| |
|
| * 1. Go to the benchmarking data page [http://data.docking.org/ data.docking.org] and select the DUD40 set, and then the [http://data.docking.org/dud40/mr/ MR set]. Download each of these five files (e.g. by right mouse clicking on them if you are in windows).
| |
|
| |
|
| [[Image:DBtut1_1f.jpg|thumb|right|Figure T1.1 - Prepare input and launch job]]
| | Step 6. Inspecting further details and poses of the ligand and decoys. |
|
| |
|
| * 2. Go to the [http://blaster.docking.org | DOCK Blaster page] and select [http://blaster.docking.org/start.shtml | Queue A Job] from the DOCK! pull down menu (Figure T1.1).
| |
|
| |
|
| * 3. Fill in the form as follows.
| | Step 7. Launching a docking job |
| ** 5. In the next field "docked ligand" select the xtal-lig.mol2 file.
| |
| ** 6. Optionally, you may select the actives.smi and inactives.smi from the next two lines.
| |
| ** 7. Enter your email address if you wish to receive progress reports by email.
| |
| ** 8. Enter a brief comment about the calculation.
| |
| ** 9. (optional: enter cofactor.par below)
| |
| | |
| If you have followed the steps above, your screen should now look something like Figure Tut1-1 (right).
| |
| | |
| | |
| ** 10. Click on "DOCK" to upload the files and begin docking. You will be taken to the "Submission Scrutinizer", as depicted in Figure Tut1-2.
| |
| | |
| | |
| ** 11. Your job should now be running. If it is not, there is either some part of the above instructions you did not follow, or we are currently experiencing problems with our systems.
| |
| | |
| ** 12. Read through the submission scrutinizer, and go to the Job Watcher by clicking on the link at the bottom of the page... http://blaster.docking.org/cgi-bin/jobmon.pl
| |
| | |
| ** 13. You should now be in the Job Watcher. Depending on how much time has elapsed, and how busy our cluster is, you will see a progress report of your job. It might look something like Figure Tut1-3. [[Image:DBtut1_3f.jpg|thumb|right|Figure T1.3]]
| |
| | |
| ** 14. If all goes well, the job will complete in about an hour. You may reload the page at anytime to get updated status of the job progress.
| |
| | |
| ** 15. Preliminary docking normally begins after the site preparation is complete, often about 15 minutes after you first submitted the job. Once preliminary docking results are available, new links appear in the job watcher to allow you to start to see docked ligands. Caution: until the preliminary docking is complete, it can be misleading to rely upon incomplete preliminary docking results to reach any conclusions about the docking results.
| |
| | |
| [[Image:DBtut1_4f.jpg|thumb|right|Figure T1.4]]
| |
| | |
| == Examining the Results ==
| |
| | |
| == Docking a Database ==
| |
| | |
| == Browsing and interpreting the results ==
| |
| | |
| == Follow up and variations ==
| |
| | |
| == Possible problems and alternative approaches ==
| |
| | |
| | |
| | |
| | |
| = Research Plan =
| |
|
| |
|
|
| |
|
| Line 77: |
Line 42: |
|
| |
|
| = Literature Cited = | | = Literature Cited = |
| | | We refer you to our publications pages for theory and practice of docking and virtual screening. |
| | | Document status: working tutorial. Please correct any errors you may find. |
| | |
| | |
| | |
| | |
|
| |
|
| [[Category:DOCK Blaster]] | | [[Category:DOCK Blaster]] |
| [[Category:Tutorials]] | | [[Category:Tutorials]] |
DOCK Blaster Tutorial 1. Dock to Human Thyroid Hormone Receptor Beta-1 (TR beta)
A DOCK Blaster Tutorial.
Step 1
a. Browse to the DOCK Blaster main page:
http://blaster.docking.org/
and select Start with a PDB code from the DOCK pull down menu:
http://blaster.docking.org/parser.shtml
b. In the PDB code field, enter 1N46, the PDB code for TR beta.
http://www.ebi.ac.uk/msd-srv/oca/oca-bin/ocashort?id=1N46
Click on DOCK to start.
If you are really impatient you can do all of the above
by simply clicking here:
http://blaster.docking.org/cgi-bin/parser.pl?code=1N46
Step 2
You will get a confirmation page showing that the target has been looked up correctly at the PDB. To proceed, click on "DOCK". Write down the DOCK Blaster Job ID number. You will need it. The docking process is now queued, and may have started if the system is not overloaded.
Step 3
Click on the link to browse progress at the bottom of the page
http://blaster.docking.org/cgi-bin/jobwatch.pl?job_id=35605&pin=
(replace the job number with your actual job number. This one will not work)
Step 4. When the job has finished, you will be in the "Calibration Done" state as shown in the progress bar with the yellow paint near the top of the page. You will see a report that reports how well docking worked.
Step 5. Interpreting the calibration summary table
Step 6. Inspecting further details and poses of the ligand and decoys.
Step 7. Launching a docking job
Literature Cited
We refer you to our publications pages for theory and practice of docking and virtual screening.
Document status: working tutorial. Please correct any errors you may find.