DOCK 3.7 tutorial based on Webinar 2017/06/28
This tuturial expands on a webinar presentation to the SBGrid [1]
Find the Youtube video here: video
Scenario 1:
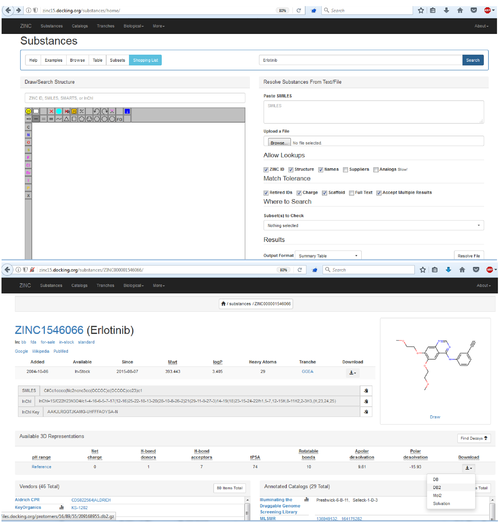
Use docking to predicted how Erlotinib (an approved drug) binds to the Epidermal Growth Factor Receptor
Get the link from the zinc webpage and us wget to download:
wget http://files.docking.org/protomers/16/89/55/209168955.db2.gz
Put the path of the downloaded database into the split database index file (this file usually contain many db2 file):
ls /path/tutorial_for_webinar/dock3.7/209168955.db2.gz > ligands.sdi
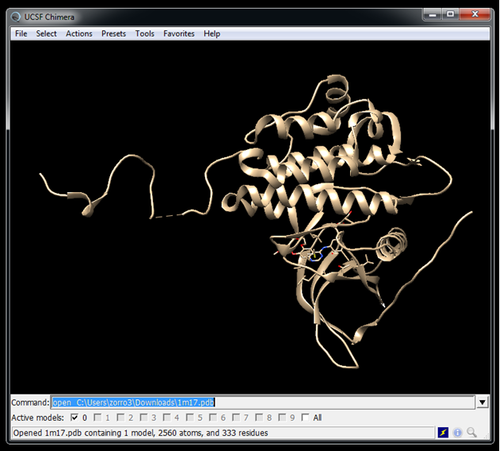
Get the receptor structure from the PDB website
wget https://files.rcsb.org/download/1M17.pdb --no-check-certificate
You may use a program like Chimera for this
Receptor file must be called: rec.pdb
Ligand file: xtal-lig.pdb
What if the crystal does not have a ligand:
Place atoms in the site were you want to dock. One way is to run sphgen and selecting spheres near residues in the site convert to pdb
Make the recptor file (remove alternative side chains):
grep "^ATOM" 1M17.pdb | grep -v ^................B > rec.pdb
Make ligand file:
grep AQ4 1M17.pdb | sed -e 's/HETATM/ATOM /g' > xtal-lig.pdb
Run blastermaster: input rec.pdb, xtal-lig.pdb and makes all receptor file need for docking.
python $DOCKBASE/proteins/blastermaster/blastermaster.py --addhOptions=" -HIS -FLIPs " -v
This command may take several minutes to run.
Here are the files that are produce:
-rw-r--r--. 1 tbalius bks 3163 Jun 17 12:27 INDOCK dockfiles/: total 30388 -rw-r--r--. 1 tbalius bks 1206051 Jun 17 12:27 ligand.desolv.heavy -rw-r--r--. 1 tbalius bks 1206051 Jun 17 12:27 ligand.desolv.hydrogen -rw-r--r--. 1 tbalius bks 3376 Jun 17 12:27 matching_spheres.sph -rw-r--r--. 1 tbalius bks 908086 Jun 17 12:27 trim.electrostatics.phi -rw-r--r--. 1 tbalius bks 3121095 Jun 17 12:27 vdw.bmp -rw-r--r--. 1 tbalius bks 1653 Jun 17 12:27 vdw.parms.amb.mindock -rw-r--r--. 1 tbalius bks 24660016 Jun 17 12:27 vdw.vdw
Modifying INDOCK File
The following parameter how much orienting to do:
match_goal 5000
Reduce 1000 if docking takes to long.
The following parameter specifies the number of poses to write out:
number_save 1 number_write 1
Consider writing out 100
Here are the minimization parameters:
# MINIMIZATION minimize no sim_itmax 500 sim_trnstep 0.2 sim_rotstep 5.0 sim_need_to_restart 1.0 sim_cnvrge 0.1 min_cut 1.0e15 iseed 777
When these parameters are turned on, dock will minimize the 6 degrees of freedom (3 rotation, 3 translation) for the poses written out. All molecules written out will be minimized.
Make prepare docking directories.
$DOCKBASE/docking/setup/setup_db2_zinc15_file_number.py ./ ligand ligand.sdi 500 count
Submit jobs to queue (we use SGE queuing system):
$DOCKBASE/docking/submit/submit.csh
To analyze the results we need to combine the results and then get poses
$DOCKBASE/analysis/extract_all.py $DOCKBASE/analysis/getposes.py
It is also possible to run dock locally:
$DOCKBASE/docking/DOCK/bin/dock64
Output: OUTDOCK and test.mol2.gz
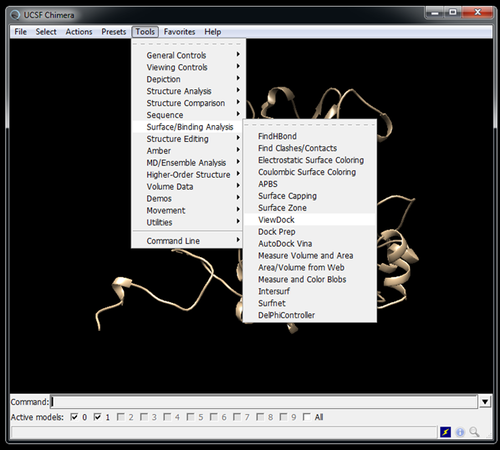
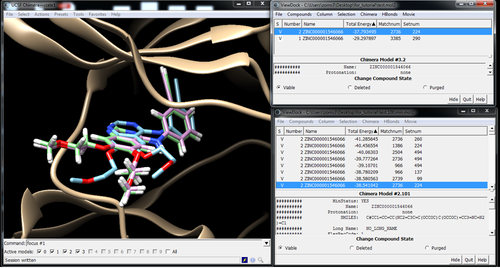
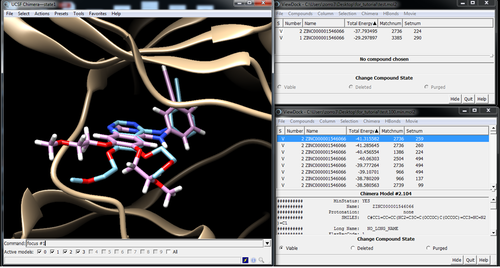
Visualize poses in Chimera with Viewdock
Pose with and without minimization: Energy -37.79 -> -38.54
Best pose out of the top 100 after min: Energy -41.31
Scenario 2:
Use docking to test enrichment capabilities of Epidermal Growth Factor Receptor using 12 ligands and DUD-E property matched decoys
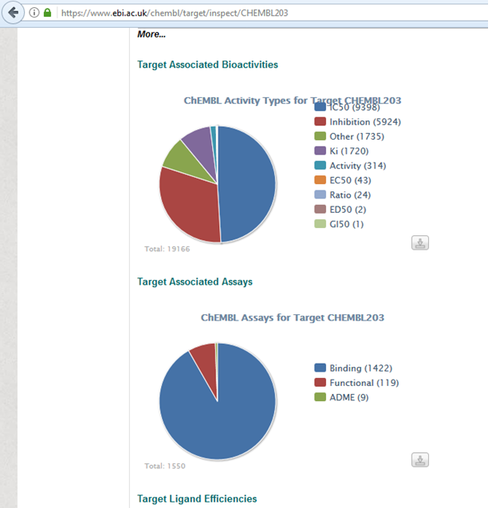
Get Known Ligands for Docking
https://www.ebi.ac.uk/chembl/target/inspect/CHEMBL203
Get Known Ligands for Docking
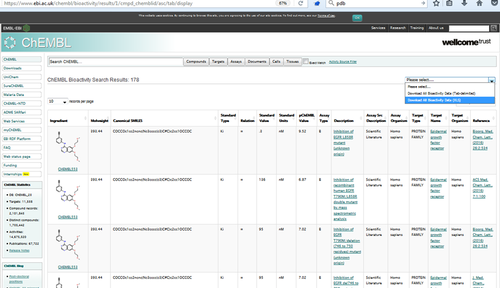
Process the XLS file into the a smiles file called "ligands_12_from_chembl.smi" with the smiles in the first column and the name in the second column.
cat bioactivity-17_20_35_00.xls | grep -v "CMPD_CHEMBLID" | head -12 | awk '{print $10 " " $1}' > ligands_12_from_chembl.smi
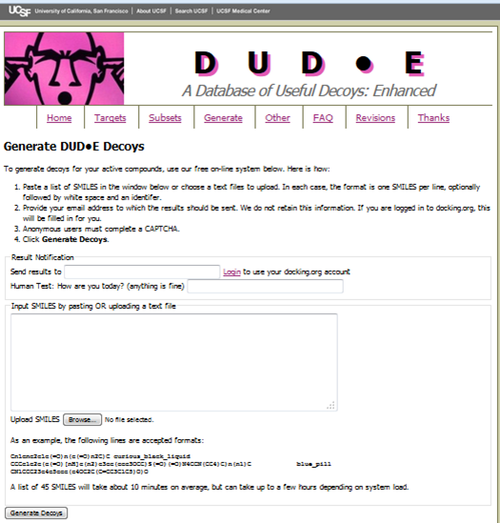
Generate Decoy Smiles File
http://dude.docking.org/generate
If the system is in DUD-E, You may download ready to dock databases here:
http://autodude.docking.org/
Here I just used the first 12 ligands from Chembl
Generated decoys using DUD-E webserver Use the link in email:
wget http://dude.docking.org/generate/results/4094969748/dude-decoys.tar.gz tar -xzvf dude-decoys.tar.gz
grep -v ligand dude-decoys/decoys/decoys.P*.picked | awk -F: '{print $2}' | awk '{print $1 " " $2}' > ! decoys.smi
${DOCKBASE}/ligand/generate/build_database_ligand.sh -H 7.4 ligands_12_from_chembl.smi
csh wraper_queue_build_smiles_ligand_mod_corina.csh decoys.smi
For more information see the page: Ligand_preparation_-_20170424
Make a list of all the databases:
ls /path/databases/ligands_12_from_chembl/CHEMBL*/*.db2.gz /path/databases/decoys/sgejob_*/finished/C*/*.db2.gz > ! ligands_decoys.sdi
awk '{print $2}' databases/ligands_12_from_chembl.smi > databases/ligands_names.txt
awk '{print $2}' databases/decoys.smi > databases/decoys_names.txt
Make directories for docking:
$DOCKBASE/docking/setup/setup_db2_zinc15_file_number.py ./ ligands_decoys databases/ligands_decoys.sdi 100 count
Submit docking jobs:
$DOCKBASE/docking/submit/submit.csh
Process results combining results and get the best poses:
$DOCKBASE/analysis/extract_all.py $DOCKBASE/analysis/getposes.py
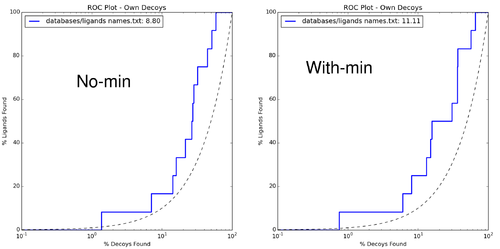
Calculate enrichments:
$DOCKBASE/analysis/enrich.py -i . -l databases/ligands_names.txt -d databases/decoys_names.txt $DOCKBASE/analysis/plots.py -i . -l databases/ligands_names.txt -d databases/decoys_names.txt