Andrii's notes on SynthI: Difference between revisions
No edit summary |
(Manual for enumeration using new code.) |
||
| Line 54: | Line 54: | ||
Analog generation doesn't seem to use similarity threshold value. Usage of large synthon library may be useful for analog generation, but speed needs to be tested. | Analog generation doesn't seem to use similarity threshold value. Usage of large synthon library may be useful for analog generation, but speed needs to be tested. | ||
== Enumeration with the updated SynthI (work in progress) == | |||
Current version of SynthI is at /mnt/nfs/exa/work/ak87/PROGRAM/SynthI-master-CC. The main changes from the original version are | |||
* support of two synthon files as input, one per each reagent | |||
* output of the synthon IDs and reaction ID, that led to a specific compound. | |||
<syntaxhighlight lang="shell"> | |||
source /nfs/soft2/anaconda3/bin/activate SynthI-env | |||
</syntaxhighlight>Prepare synthons for a specific reaction. List of supported reactions can be found in the .pdf file in the parent dir. First, make .smi file with SMILES and id of each building block (e.g. amine and carboxylic acid). Then prepare synthons from each of the BB files:<syntaxhighlight lang="shell"> | |||
python /mnt/nfs/exa/work/ak87/PROGRAM/SynthI-master-CC/SynthI_BBsBulkClassificationAndSynthonization.py -i bb-list.smi | |||
</syntaxhighlight>Leave only SMILES and names<syntaxhighlight lang="shell"> | |||
awk '{print $1 " " $NF}' bb_list.smi_Synthmode.smi > synth.smi | |||
</syntaxhighlight>Then you can enumerate the library based on two reagent (synthon) lists:<syntaxhighlight lang="shell"> | |||
python /mnt/nfs/exa/work/ak87/PROGRAM/SynthI-master-CC/SynthI_BulkFragmentationEnumerationAndAnaloguesDesign.py -i synthon_X.smi -i2 synthon_Y.smi -oD RESULTS/ --enumerationMode --nCores 2 | |||
</syntaxhighlight>You will get results in RESULTS/FinalOut_allEnumeratedCompounds_DuplicatesCanBePresent.smi: | |||
C=C(C)C(O)(C(=O)n1cc(C(C)=O)cc1C(=O)NCc1ccccc1)C(F)(F)F R5.3_CSSB00155635782_CSSB00000019219_ | |||
Where R5.3 is the reaction ID (see .pdf), followed by two synthon IDs. The script is also capable of invoking only specified reactions, by using flags <code>--fragmentationMode include_only --reactionsToWorkWith "R3, R5"</code> (not tested). | |||
=TODO= | =TODO= | ||
# Work with 2 synthon files, instead of one. | # Work with 2 synthon files, instead of one. -- DONE | ||
# Output identifiers of enumerated compds: RXN_synt1_synt2 | # Output identifiers of enumerated compds: RXN_synt1_synt2 -- DONE | ||
# Processing the end of synthon file | # Processing the end of synthon file | ||
Revision as of 02:48, 22 March 2022
Parent page: SynthI
Preparing analogs with SynthI
Working with https://cartblanche22.docking.org/searchZinc/ZINCoT000006Aq87, bash needed. Prepare .smi file with the list of SMILES (and names) of compds to prepare analogs for.
First, we need to fragment our compounds:
python /nfs/soft2/SynthI//SynthI_BulkFragmentationEnumerationAndAnaloguesDesign.py -i ligand.smi --nCores 5 --MaxNumberOfStages 1
In the file ligand.smi_out you will get a list of synthons and reactions that are applied to the molecule:
C[C@@H](NCc1cccc(-n2cccn2)c1)c1csc2ccccc12 ZINCoT000006Aq87 c1cn[nH:20]c1.c1cc(C[NH2:20])c[cH:21]c1.C[CH2:10]c1csc2ccccc12 R3.1_0|R5.2_0 3 0 AvailableSynthons: NotAvailableSynthons:C[CH2:10]c1csc2ccccc12|c1cc(C[NH2:20])c[cH:21]c1|c1cn[nH:20]c1
As the synthons generated contain molecular fragments, you will have to manually cap the BBs according to the reactions provided. Then search for similar BBs in SmallWorld. For each of the found lists of BBs do:
awk '{print $1 " " $2}' thioph-Cl.tsv | grep -v alignment > thioph-Cl.smi
Then cat into one file and prepare synthons from the BBs found.
python /nfs/soft2/SynthI/SynthI_BBsBulkClassificationAndSynthonization.py -i bb_analogs.smi
Leave only SMILES and names
awk '{print $1 " " $NF}' bb_analogs.smi_Synthmode.smi > bb_analogs_synth.smi
A. Enumeration based on all found BBs
Directory under "-oD" will contain eitherFinalOut_allEnumeratedCompounds_DuplicatesCanBePresent.smi or AnalogsForMol1.smi file with list of SMILES for generated compds.
Different from analog generation, the output of enumeration does not contain the reactions and synthons used for compod. generation.
Using all available reactions:
mkdr ENUMERATED python /nfs/soft2/SynthI/SynthI_BulkFragmentationEnumerationAndAnaloguesDesign.py -i bb_analogs_synth.smi --nCores 10 -oD ENUMERATED --MaxNumberOfStages 5 --enumerationMode --MWupperTh 460 --MWlowerTh 200
Morgan2 Tc of obtained compds to the parent.
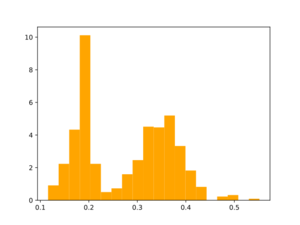
Using the same reactions that were used for initial fragmentation:
mkdir ENUMERATED-R3-R5 python /nfs/soft2/SynthI/SynthI_BulkFragmentationEnumerationAndAnaloguesDesign.py -i bb_analogs_synth.smi --nCores 10 -oD ENUMERATED-R3-R5/ --MaxNumberOfStages 5 --enumerationMode --MWupperTh 460 --MWlowerTh 200 --fragmentationMode include_only --reactionsToWorkWith "R3, R5"
Morgan2 Tc of obtained compds to the parent.
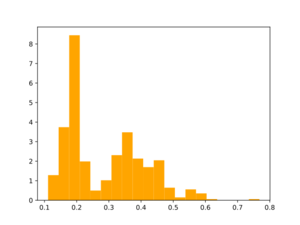
B. Analogs from a synthon library
python /nfs/soft2/SynthI/SynthI_BulkFragmentationEnumerationAndAnaloguesDesign.py -i ligand.smi --SynthLibrary bb_analogs_synth.smi --simTh 0.5 --analoguesLibGen --nCores 10 -oD ANALOGS --MaxNumberOfStages 5 --desiredNumberOfNewMols 1000 --enumerationMode --MWupperTh 460 --MWlowerTh 200
Morgan2 Tc of obtained compds to the parent.
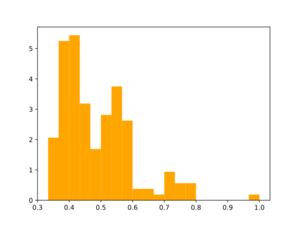
Analog generation doesn't seem to use similarity threshold value. Usage of large synthon library may be useful for analog generation, but speed needs to be tested.
Enumeration with the updated SynthI (work in progress)
Current version of SynthI is at /mnt/nfs/exa/work/ak87/PROGRAM/SynthI-master-CC. The main changes from the original version are
- support of two synthon files as input, one per each reagent
- output of the synthon IDs and reaction ID, that led to a specific compound.
source /nfs/soft2/anaconda3/bin/activate SynthI-env
Prepare synthons for a specific reaction. List of supported reactions can be found in the .pdf file in the parent dir. First, make .smi file with SMILES and id of each building block (e.g. amine and carboxylic acid). Then prepare synthons from each of the BB files:
python /mnt/nfs/exa/work/ak87/PROGRAM/SynthI-master-CC/SynthI_BBsBulkClassificationAndSynthonization.py -i bb-list.smi
Leave only SMILES and names
awk '{print $1 " " $NF}' bb_list.smi_Synthmode.smi > synth.smi
Then you can enumerate the library based on two reagent (synthon) lists:
python /mnt/nfs/exa/work/ak87/PROGRAM/SynthI-master-CC/SynthI_BulkFragmentationEnumerationAndAnaloguesDesign.py -i synthon_X.smi -i2 synthon_Y.smi -oD RESULTS/ --enumerationMode --nCores 2
You will get results in RESULTS/FinalOut_allEnumeratedCompounds_DuplicatesCanBePresent.smi:
C=C(C)C(O)(C(=O)n1cc(C(C)=O)cc1C(=O)NCc1ccccc1)C(F)(F)F R5.3_CSSB00155635782_CSSB00000019219_
Where R5.3 is the reaction ID (see .pdf), followed by two synthon IDs. The script is also capable of invoking only specified reactions, by using flags --fragmentationMode include_only --reactionsToWorkWith "R3, R5" (not tested).
TODO
- Work with 2 synthon files, instead of one. -- DONE
- Output identifiers of enumerated compds: RXN_synt1_synt2 -- DONE
- Processing the end of synthon file