Search results
Jump to navigation
Jump to search
Page title matches
- USAGE: grid -i grid.in [-stv] [-o grid.out] - i input_file #Input parameters extracted from input_file, or grid.in if not specified10 KB (1,588 words) - 20:25, 8 October 2012
- GRID-CONVERT AUTHOR: Kaushik Raha, John J. Irwin (Derived from Todd Ewing's GRID Program)485 bytes (65 words) - 01:02, 11 March 2014
- ...s based on the non-bonded terms of the molecular mechanic force field (see Grid for more background). Grid Score Parameters1 KB (189 words) - 17:28, 15 February 2014
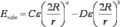
File:Grid vdw term.jpg (312 × 70 (6 KB)) - 00:44, 12 March 2014File:Grid score function.JPG (322 × 76 (7 KB)) - 00:44, 12 March 2014- ALL ABOUT SGE (SUN GRID ENGINE) # when the value is empty or path does not exit on the system, Grid Engine25 KB (3,771 words) - 20:56, 23 January 2017
- * MakeDOCK automates the process of sphere and grid generation for a target. * Automatically displays non-fatal WARNINGS that occurred during grid generation, which often lead to incorrect docking results.3 KB (488 words) - 02:43, 13 March 2014
- Grid inhomogeneous Solvation Theory (GIST) is a method for calculating thermodyn get center of grid from aligned.lig.pdb26 KB (4,023 words) - 00:21, 5 March 2019
- Grid inhomogeneous Solvation Theory (GIST) is a method for calculating thermodyn ...in without any cofactors [[Tutorial on running Molecular Dynamics for GIST grid generation with scripts 2]].49 KB (7,168 words) - 00:18, 9 November 2017
- 5 KB (793 words) - 16:26, 3 May 2017
Page text matches
- GRID-CONVERT AUTHOR: Kaushik Raha, John J. Irwin (Derived from Todd Ewing's GRID Program)485 bytes (65 words) - 01:02, 11 March 2014
- ...s based on the non-bonded terms of the molecular mechanic force field (see Grid for more background). Grid Score Parameters1 KB (189 words) - 17:28, 15 February 2014
- ...gly refer to this as "SGE", but that is the name of an older product - Sun Grid Engine - that we no longer use.664 bytes (107 words) - 16:51, 3 June 2015
- ...ce (see Grid). Atom VDW overlaps are penalized by checking the bump filter grid, or with the contact_clash_overlap parameter for the intramolecular score. o contact_score_grid_prefix [grid] (string):2 KB (235 words) - 18:10, 8 October 2012
- ...ation. Docktools consists of chemgrid, solvmap, solvgrid, grid-convert and grid-convrds. This section will briefly review the theory behind each of the pr732 bytes (106 words) - 20:11, 8 October 2012
- #REDIRECT [[Understanding MakeDOCK, which automates sphere and grid generation]]80 bytes (9 words) - 20:33, 8 October 2012
- Grid Engine job queuing and management software.2 members (0 subcategories, 0 files) - 16:52, 3 June 2015
- 1.1) The VDW grid 1.2) The electrostatics grid909 bytes (142 words) - 00:26, 25 May 2024
- * [[Tutorial on running Molecular Dynamics for GIST grid generation]] * [[Tutorial on running Molecular Dynamics for GIST grid generation with scripts]]437 bytes (61 words) - 19:56, 15 November 2017
- ...amount of atom VDW overlap is defined with the bump_overlap parameter (see Grid). At the time of bump evaluation the number of allowed bumps is defined wit * bump_grid_prefix [grid] (string):1 KB (210 words) - 18:10, 8 October 2012
- the grid generation scripts all start with do_*.csh, i.e. do_chemgrid.csh the other grid generation.635 bytes (110 words) - 03:39, 14 February 2014
- ...ed high dielectric volume displaced by the protein that is stored for each grid element in the active site. Thus the volume based ligand desolvation energy ...nd complete exposure to the solvent on the protein surface. However, being grid-based it is fast and can be used during conformational search and final sco2 KB (278 words) - 00:26, 25 May 2024
- ...gand interactions. J. Mol. Biol. 161, 269-288 (1982)]</ref>. The program [[grid]] ....1002/jcc.540130412 Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. Journal of Computational Chemistry 13, 505-524 (192 KB (229 words) - 22:43, 10 January 2019
- We use one of several OpenSGEs - open versions of sun grid engine, as a queuing system.178 bytes (28 words) - 18:11, 22 September 2014
- 0.4 ; grid spacing in angstroms output_prefix ; output grid prefix name3 KB (436 words) - 17:44, 15 February 2014
- ...ontinuous scoring is requested, then score parameters normally supplied to grid, must also be supplied to DOCK. It is left to the user to make sure consist #Flag to perform continuous non-grid based scoring as the primary3 KB (343 words) - 18:46, 15 February 2014
- * ligand.desolv.heavy - ligand desolvation scoring grid, non-H atoms * ligand.desolv.hydrogen - ligand desolvation scoring grid, H atoms2 KB (294 words) - 15:10, 25 March 2014
- DOCK3.5 score is a variant of Grid-based scoring (see Grid). DOCK3.5 score function calculates ligand desolvation in addition to steri #Calculate electrostatic interaction from ESP grid calculated using DelPhi3 KB (337 words) - 20:02, 8 October 2012
- ...ligand atom. For the receptor, these can be precalculated and stored on a grid. øDES,BULK(xi) and øDES,EXPL(xi) are interpolated from grids calculated ...vely. Bulk and explicit desolvation grids are calculated from fj and Sj at grid points p, distance rjp from the receptor atom multiplied by gaussian weight3 KB (475 words) - 00:26, 25 May 2024
- ...stored on grids. The calculations are based on the theory presented in the Grid section above. However only the steric interaction energy grids are used in 0.33 ; grid spacing in angstroms3 KB (365 words) - 05:24, 13 March 2014