DOCK 3.7 2015/04/15 abl1 Tutorial
This tutorial use the 3.7.2 beta version of dock release on April 17, 2015.
This is for a Linux environment and the scripts assume that you are running on SGE queuing system.
set up directories and get databases
Create directory called "RotationProject"
create a python file called "autodude_db_download.py"
# this gets the database from the autodude webpage import sys, os import urllib system = 'abl1' url = 'http://autodude.docking.org/dude_e_db2/' print "url = " + url #page=requests.get(url) webfile = urllib.urlopen(url) page = webfile.read() webfile.close() splitpage=page.split('\n') for line in splitpage: if system in line: file = line.replace('"',' ').split()[2] print url+file urllib.urlretrieve(url+file,file) # exit()
This python script will download the dockable db2 databases from the autodude webpage.
python /mnt/nfs/home/rstein/RotationProject/autodude_db_download.py
make a subdirectory called databases:
mkdir databases
go inside.
cd databases
make directories for ligands and decoys and move the corresponding files into those directories
mkdir decoys mv decoys*db2.gz decoys
mkdir ligands mv ligands*db2.gz ligands
download the ligand and decoy isomeric smiles file:
wget http://autodude.docking.org/abl1/decoys_final.ism mv decoys_final.ism decoys.ism
note that the scripts expect the name to be decoys.ism, so we changed the name.
wget http://autodude.docking.org/abl1/actives_final.ism mv actives_final.ism ligands.ism
run be_blasti.py
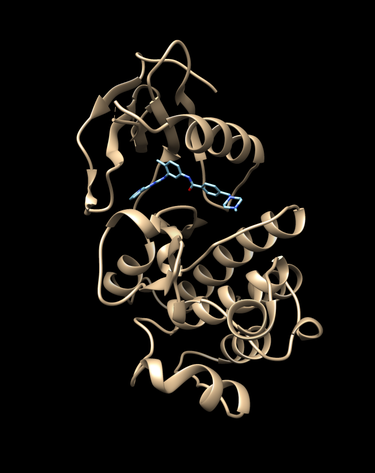
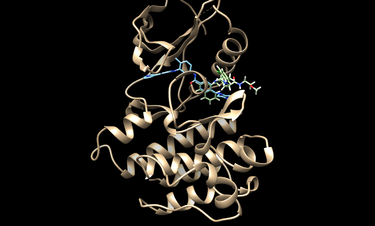
First we need to get our protein of interest from the protein databank (pdb). We will typiclly use a receptor with a ligand bound as is the case for pdbcode 2HYY, which is the Abl kinase domain in complex with imatinib (STI571, Glivec).
Note, in the following scripts, that DOCKBASE is a environment variable that point to the DOCK3.7 code. e.g.:
setenv DOCKBASE "/path2dock3.7/DOCK"
or
setenv DOCKBASE "/nfs/home/tbalius/zzz.github/DOCK"
Note, if you get an error ImportError: No module named Bio.PDB, then install the biopython as followed:
sudo yum install python-biopython
Create the following cshell script 0001.be_balsti_py.csh by using your favorite text editor (eg vim).
Note that the "#" symbol denotes a comment to explain what the script is doing.
#!/bin/csh
# this script calls be_blasti.py which creates a receptor and ligand file from a (list of) pdbcode(s).
# msms is a molecular surface generation program needed for be_blasti.py to run
# which is put in your path
set path = ( /nfs/home/tbalius/zzz.programs/msms $path )
# you will need to have msms on you system.
set list = "2HYY" # or use `cat filename` to list your pdb codes here from a text file like pdblist_rat, to loop over each variable (pdb code) later
#set list = `cat $1`
#set list = `cat /nfs/work/users/tbalius/VDR/Enrichment/pdblist_rat `
# CHANGE THIS, according to where the magic is going to happen
#set mountdir = "/mnt/nfs/work/users/tbalius/VDR/"
set mountdir = `pwd`
# loop over pdbnames e.g. 1DB1 or list
foreach pdbname ( $list )
echo " ${pdbname} "
# for each pdb makes a directory with its name
set workdir = ${mountdir}/${pdbname}
## so you don't blow away stuff; continue means STOP here and continue with next pdb from list
if ( -s $workdir ) then
echo "$workdir exits"
continue
endif
mkdir -p ${workdir}
cd ${workdir}
# the atom type definition is needed for msms which is sym-linked into the cwd
ln -s /nfs/home/tbalius/zzz.programs/msms/atmtypenumbers .
# carbs are disregarded as ligands! if it is: carbohydrate instead of noncarbohydrate
# renumber renumbers the residue number
python $DOCKBASE/proteins/pdb_breaker/be_blasti.py --pdbcode $pdbname nocarbohydrate original_numbers | tee -a pdbinfo_using_biopython.log
# error checking looks for receptor and ligand file which should be produced by be_blasti.py
if !(-s rec.pdb) then
echo "rec.pdb is not found"
endif
mv rec.pdb temp.pdb
grep -v TER temp.pdb | grep -v END > rec.pdb
rm temp.pdb
# be_blasti.py produces peptide which may be used as a ligand if no other ligand is produced
if (-s lig.pdb) then
sed -e "s/HETATM/ATOM /g" lig.pdb > xtal-lig.pdb
else if (-s pep.pdb) then ## if no ligand and peptide
sed -e "s/HETATM/ATOM /g" pep.pdb > xtal-lig.pdb
else
echo "Warning: No ligand or peptid."
endif
end # system
running 0001.be_balsti_py.csh will run a script that comes with dock called be_blasti.
issue the following command to run the script:
csh 0001.be_balsti_py.csh
It will do the following
- Download the pdb file from the web
- Break the file into rec and ligand components
Note that you will need to have msms on you system.
For Shoichet lab members msms is already installed.
check to make sure that the right ligand was selected and the the residue is not missing anything of importance. If this automatic procedure has not prepared these files correctly, then modify them.
Visualize them with chimera or an alternive visualization program like pymol.
cd 2HYY
chimera rec.pdb lig.pdb
run blastermaster.py
Write (paste what follows) the following script using a text editor like vi. This script creates the files necessary for docking including the spheres (for orienting the ligands/decoys) and grids (for scoring the ligand/decoy poses)
WARNING: if you copy and pasted the script make sure there is no space before the "EOF"; this is because "EOF" designates the "end of file" for the cat command, if a space is there it wont stop cat'ing.
0002.blastermaster.csh
#!/bin/csh
# This script runs Ryan's blastermaster python masterscript for generating everything that dock needs, i.e. grids, spheres
# Run on sgehead as jobs are submitted to the queue
# list is same as in 001... script
set list = "2HYY"
#set list = `cat $1`
#set list = `cat /nfs/work/users/tbalius/VDR/Enrichment/pdblist_all `
set mountdir = `pwd`
#set mountdir = "/nfs/work/users/tbalius/VDR/"
# loop over all pdb(s)
foreach pdbname ( $list )
echo "${pdbname}"
set workdir = ${mountdir}/${pdbname}
# checks that 001 ran successfully and produced the directory structure as expected
# if not stops with current pdb code and continues with next one in list
if ! ( -s $workdir ) then
echo "$workdir does not exit"
continue
endif
cd $workdir
#cat xtal-lig_ori.pdb | awk '{if ($1 == "ATOM" || $1 == "HETATM"){print $0}}' | sed -e "s/HETATM/ATOM /g" > xtal-lig.pdb
# the following lines create a qsub script which submits blastermaster to the queue
cat <<EOF > qsub.csh
#!/bin/csh
#\$ -cwd
#\$ -j yes
#\$ -o stderr
#\$ -q all.q
cd $workdir
python $DOCKBASE/proteins/blastermaster/blastermaster.py --addhOptions=" -HIS -FLIPs " -v
EOF
qsub qsub.csh
end # pdbname
# going to the next pdb
# this will produce two directories:
# 1) working - contains all input and output files that are generated; not needed afterwards but as a reference
# 2) dockfiles - contains everything that is needed to run dock (copied from working)
# grids
# trim.electrostatics.phi
# vdw.vdw
# vdw.bmp
# ligand.desolv.heavy
# ligand.desolv.hydrogen
# spheres
# matching_spheres.sph
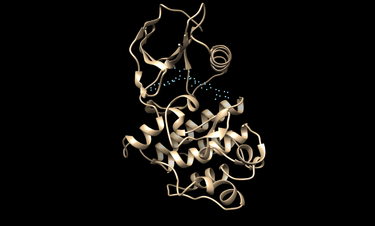
Visualize the docking spheres
Use the showsphere program:
$DOCKBASE/proteins/showsphere/bin/showsphere
$DOCKBASE/proteins/showsphere/doshowsph.csh file.sph 1 file.pdb
Alternatively, use the following simple cshell/awk script to convert spheres to pdb format:
cat << EOF > sphere2pdb.csh
#!/bin/csh -f
awk '$0!~/e/{ \
printf("ATOM %5d C SPH%5d%12.3f%8.3f%8.3f%6.2f%6.2f\nTER\n", \$1, \$1, \$2, \$3, \$4, 1, \$5)}' \$1
EOF
This command statement will write the commands script to a file called sphere2pdb.csh. Make sure there is not space before the second EOF (end of file) above. To run the command:
csh sphere2pdb.csh file.sph > file.pdb
cd 2HYY/working
chimera rec.pdb matching_spheres.pdb
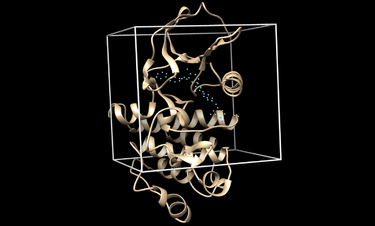
The box used for scoring can also be visualized in chimera with the following command:
chimera rec.pdb matching_spheres.pdb box
run enrichment calculations
See the following tutorial for updated scripts: DOCK_3.7_2016/09/16_Tutorial_for_Enrichment_Calculations_(Trent_&_Jiankun)
Here is the new recomend script that uses the preexisting submission infrastructure of DOCK:
The script at the following location submits a single job as an array job, ie one script is submitted, but many task are run, where each task is a separate chunk:
The following script has advantages in that it is a very intuitive script -- every db2 chunk has a qsub script that is submitted to the queue.
If you decide not to use the methods linked to above then write a file called 0003.lig-decoy_enrichment.csh
#!/bin/csh
#This script docks a DUD-e like ligand-decoy-database to evaluate the enrichment performance of actives over decoys
#It assumes that ligands and decoys have been pre-prepation (see script blablabla_ToDo) which needs to be run in SF.
# filedir is where your rec.pdb and xtal-lig.pdb and dockfiles directory live
set filedir = "/mnt/nfs/home/rstein/RotationProject" #CHANGE THIS
# this is where the work is done:
set mountdir = $filedir # Might CHANGE THIS
set dude_dir = "/mnt/nfs/home/rstein/RotationProject/databases" # should contain decoy.smi and ligand.smi for ROC script 00005...csh
## TO DO - rename this outside in the dir structure and call in blbalbalbabla script
if (-s $dude_dir) then
echo " $dude_dir exist"
else
# this is something to modified in future.
# probably better to exit if it is not there.
echo "databases do not exist. "
echo "consider making a symbolic link to the database files"
#echo "making a symbolic link:"
#echo "ln -s /mnt/nfs/work/users/fischer/VDR/27Jan2014_learningDOCKrgc/databases_all_xtal-ligand_decoy $dude_dir"
#ln -s /mnt/nfs/work/users/fischer/VDR/27Jan2014_learningDOCKrgc/databases_all_xtal-ligand_decoy $dude_dir
endif
# change if you want to use a different or consistent dock version
set dock = ${DOCKBASE}/docking/DOCK/bin/dock64
set list = "2HYY"
#set list = `cat $1`
#set list = `cat file`
# CHANGE THIS (pdbname)
foreach pdbname ( $list )
# creates "ligands" and "decoys" and has the aim to dock all of the subsets for those two
foreach db_type ( "ligands" "decoys" )
set workdir1 = "${mountdir}/${pdbname}/ligands-decoys/${db_type}"
mkdir -p ${workdir1}
cd ${workdir1}
# puts dockfiles in the right relative-path that INDOCK file expects
ln -s $filedir/${pdbname}/dockfiles .
set count = '1'
# loop over database files to put each into a seperate chunk
foreach dbfile (`ls $dude_dir/${db_type}/${db_type}*.db2.gz`)
echo $dbfile
set chunk = "chunk$count"
set workdir2 = ${workdir1}/$chunk
## so you don't blow away stuff
if ( -s $workdir2 ) then
echo "$workdir2 exits"
continue
endif
#rm -rf ${workdir}
mkdir -p ${workdir2}
cd ${workdir2}
# copy INDOCK file of choice in right location
#cp $filedir/zzz.dock3_input/INDOCK .
#cp $filedir/INDOCK_match20K INDOCK
#cp $filedir/INDOCK_5k_TolerantClash INDOCK # CHANGE THIS
cp $filedir/${pdbname}/INDOCK .
# modified the dock file using sed. here we change some key sampling parameters; sed -i changes input file internally (overwrites), -e changes file externally (pipes it to screen or into file if redirected)
#sed -i "s/bump_maximum 50.0/bump_maximum 500.0/g" INDOCK
#sed -i "s/bump_rigid 50.0/bump_rigid 500.0/g" INDOCK
#sed -i "s/check_clashes yes/check_clashes no/g" INDOCK
ln -s $dbfile .
set dbf = `ls *.gz`
echo "./$dbf"
# says what to dock and where it sits
echo "./$dbf" > split_database_index
# writes submission script that runs dock on the sgehead queue
cat <<EOF > DOCKING_${db_type}.csh
#\$ -S /bin/csh
#\$ -cwd
#\$ -q all.q
#\$ -o stdout
#\$ -e stderr
cd ${workdir2}
echo "starting . . ."
date
echo $dock
$dock
date
echo "finished . . ."
EOF
qsub DOCKING_${db_type}.csh
# alternatively if you don't want to run it on the queue but locally comment in this instead:
#csh DOCKING_${lig_type}.csh &
@ count = ${count} + 1
# counter is chuch dir
end # dbfile
end # db_type
end # pdbname
combine scores and poses
Write this file as 0004.combineScoresAndPoses.csh. This script combines your docking runs (for ligands and decoys) and stores them in the extract_all.txt file. It also creates a .mol2 file containing all top scoring poses for each ligand/decoy.
#!/bin/csh
# This script combines the results from the ligand-decoy run 0003 (all chunks) into a combine file containing dock scores from OUTDOCK files
# Three files are produced (one for lig, decoy and both)
# and: a file which has top poses as specified (e.g. top 1000 molecules with 2 poses each); two files (for lig and for decoys)
# to remove dir
# rm -fr pdbs/3O1D/ligands-decoys/ligands/allChunksCombined/ pdbs/3O1D/ligands-decoys/decoys/allChunksCombined/ pdbs/3O1D/ligands-decoys/dockedLigDecoyCombined/
set filedir = "/mnt/nfs/home/rstein/RotationProject"
set mountdir = "/mnt/nfs/home/rstein/RotationProject"
set d37 = $DOCKBASE/analysis/
cd $mountdir
set list = "2HYY"
#set list = `cat filename`
#set list = `cat $1`
foreach pdbname ( $list )
foreach db_type ( "ligands" "decoys" )
set workdir = ${mountdir}/${pdbname}/ligands-decoys/${db_type}/allChunksCombined
echo $pdbname
#ls -l ${mountdir}/${pdbname}/${db_type}/
mkdir -p ${workdir}
cd ${workdir}
# creates a file called dirlist that contains the full path of all directories with docked runs (chunks)
ls -ld ${mountdir}/${pdbname}/ligands-decoys/${db_type}/* | awk '/chunk/{print $9}' > dirlist
#ls -ld ${mountdir}/${pdbname}/ligands-decoys/${db_type}/*
# for debuging
#echo "print $db_type dirlist:"
#cat dirlist
# script extracts scores from all docking runs specified in dirlist
$d37/extract_all.py
# script gets poses for top scoring molecules and produces poses.mol2 (default name)
$d37/getposes.py -d ${mountdir}/${pdbname}/ligands-decoys/${db_type}
end # db_type
## combine decoyes and actives
set workdir = ${mountdir}/${pdbname}/ligands-decoys/dockedLigDecoyCombined
rm -rf ${workdir}
mkdir -p ${workdir}
cd ${workdir}
cat ${mountdir}/${pdbname}/ligands-decoys/ligands/allChunksCombined/dirlist ${mountdir}/${pdbname}/ligands-decoys/decoys/allChunksCombined/dirlist > dirlist
# for debuging
#echo "print ALL dirlist"
#cat dirlist
$d37/extract_all.py
#$d37/getposes.py -d ${mountdir}/${pdbname} # doesn't work yet; not really needed
#getposes.py -z -l 1000 -x 2 -f extract_all.sort.uniq.txt -o ligands.1000.mol2 -d /mnt/nfs/work/users/fischer/VDR/27Jan2014_learningDOCKrgc/Enrichment/1DB1/DOCKING/ligands
end # pdbname
create AUC plot of ligands and decoys
Make sure that you are using a version of python that has matplotlib/numpy/scipy modules:
For Shoichet user source the following:
source /nfs/soft/python/envs/complete/latest/env.sh
or
source /nfs/soft/python/envs/complete/latest/env.csh
Also, if you are running the processing script remotely make sure to use X11 forwarding by including a -X in the ssh command. eg:
ssh gimel.ucsf.bkslab.org -X
Write a file called 0005.AUCplot_of-lig-decoys.csh.
#!/bin/csh
# This script creates a log adjusted AUC (ROC) plot with ligand vs decoy results
# need X11 forwarding enabled when running remotely (ssh sgehead -X).
set filedir = "/mnt/nfs/home/rstein/RotationProject" #CHANGE THIS
set mountdir = "/mnt/nfs/home/rstein/RotationProject" #CHANGE THIS
set d37 = $DOCKBASE/analysis
set dude_dir = "/mnt/nfs/home/rstein/RotationProject/databases" # should contain decoy.smi and ligands.smi
# ln -s /mnt/nfs/work/users/fischer/VDR/lig-decoy-db/ligands.mod.smi /mnt/nfs/work/users/fischer/VDR/lig-decoy-db/ligands.smi
# CHANGE THIS
set list = "2HYY"
#set list = `cat filename`
#set list = `cat $1`
foreach pdbname ( $list )
set workdir = ${mountdir}/${pdbname}/ROC_ligdecoy/
# This script will not work without the following line:
echo "HERE is the HAWK"
# checks that previous script 0003 has produced mol2 files
if (! ( -s $mountdir/${pdbname}/ligands-decoys/decoys/allChunksCombined/poses.mol2) && ! (-s $mountdir/${pdbname}/ligands-decoys/ligands/allChunksCombined/poses.mol2 )) then
ls -l $mountdir/${pdbname}/ligands-decoys/decoys/allChunksCombined/poses.mol2
ls -l $mountdir/${pdbname}/ligands-decoys/ligands/allChunksCombined/poses.mol2
echo "skipping ${pdbname}. cannot generate ROC"
continue
endif
rm -rf $workdir
mkdir -p $workdir
cd $workdir
#wget http://dude.docking.org/targets/aa2ar/actives_final.ism
# reads ZINC ids (ligand or decoy molecule names)
# everything
awk '{print $2}' $dude_dir/decoys.ism > decoys.name # note that you may have to change the column ($2) based on where the SMILES codes are
awk '{printf "%9s\n", $3}' $dude_dir/ligands.ism > ligands.name # note that you may have to change the column ($3) based on where the SMILES codes are
#things that finished docking
awk '{print $3}' $mountdir/${pdbname}/ligands-decoys/decoys/allChunksCombined/extract_all.sort.uniq.txt > decoys.finished.name
awk '{print $3}' $mountdir/${pdbname}/ligands-decoys/ligands/allChunksCombined/extract_all.sort.uniq.txt > ligands.finished.name
cat ${mountdir}/${pdbname}/ligands-decoys/ligands/allChunksCombined/dirlist ${mountdir}/${pdbname}/ligands-decoys/decoys/allChunksCombined/dirlist > dirlist
#which enrich.py
set enrich_py = $d37/enrich.py
set plots_py = $d37/plots.py
pwd
# calculates AUCs, stores in txt file which is then plotted for finished ligands and decoys
python ${enrich_py} -i . -o . --ligand-file=ligands.finished.name --decoy-file=decoys.finished.name
python ${plots_py} -i . -o . --ligand-file=ligands.finished.name --decoy-file=decoys.finished.name -l $pdbname
mv roc.txt roc.finished.txt
mv roc_own.txt roc_own.finished.txt
mv roc_own.png roc_own.finished.png
#
# calculates AUCs, stores in txt file which is then plotted for all ligands and decoys
# - i is the flag for the input directory, this dir should contain the extract_all.sort.uniq.txt.
# the scripts enrich_py and plots_py will go through the extract file and look for the ligand and decoy names.
# when it finds them it will populate the ROC cruve. these values are devied by the total number of ligand or decoys.
# note that often not all ligands and not all decoys finish so the point (1,1) is always included and interpolations is performed . . .
#
#python ${enrich_py} -i $mountdir/${pdbname}/ligands-decoys/dockedLigDecoyCombined/ -o . --ligand-file=ligands.name --decoy-file=decoys.name
#python ${plots_py} -i $mountdir/${pdbname}/ligands-decoys/dockedLigDecoyCombined/ -o . --ligand-file=ligands.name --decoy-file=decoys.name -l $pdbname
python ${enrich_py} -i . -o . --ligand-file=ligands.name --decoy-file=decoys.name
python ${plots_py} -i . -o . --ligand-file=ligands.name --decoy-file=decoys.name -l $pdbname
end #pbdname
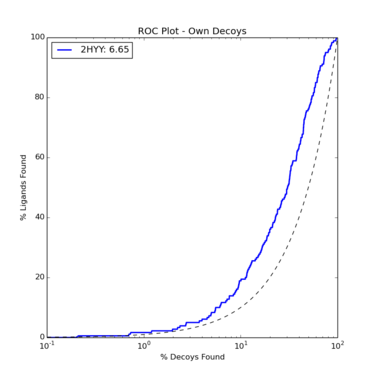
The ROC plot for ligands and decoys docking to Abl1 looks like this:
This shows poor enrichment, though it is better than random (indicated by the dotted line). This is to be expected as docking to kinases is notoriously difficult. Two changes to our procedure would be to delete some of the spheres to focus docking on a particular region of the protein we are most interested in, as well as increasing the polarity of the hinge region of Abl1 to allow for more hydrogen bonding.
improving docking
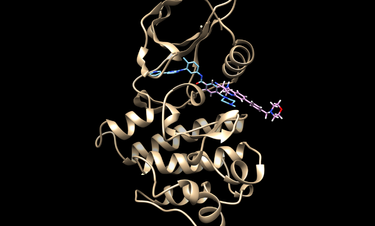
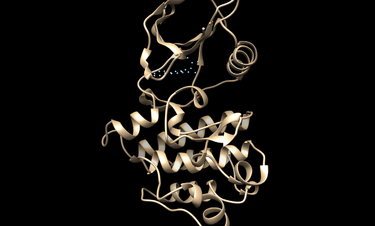
Modify matching spheres
The docking could be potentially improved by modifying the spheres and orienting the ligands/decoys to only those spheres within the hinge region of Abl1. The following image show modified spheres:
To modify spheres: first, convert them to pdb format; then, visualize them in your favorite program (pymol, chimera, etc.); then, delete, move, or add atoms to this file; and finally convert it back to the sph format.
As discussed above [[1]], you may use doshowsph.csh to convert the spheres to pdb format.
Here is the script that will convert a pdb file into a sphere file.
$DOCKBASE/proteins/pdbtosph/bin/pdbtosph
The 0003.lig-decoy_enrichment.csh would be modified to include this line and renamed to 0003.lig-decoy_enrichment_mod_sph.csh:
!/bin/csh
#This script docks a DUD-e like ligand-decoy-database to evaluate the enrichment performance of actives over decoys
#It assumes that ligands and decoys have been pre-prepation (see script blablabla_ToDo) which needs to be run in SF.
# filedir is where your rec.pdb and xtal-lig.pdb and dockfiles directory live
set filedir = "/mnt/nfs/home/rstein/RotationProject" #CHANGE THIS
# this is where the work is done:
set mountdir = $filedir # Might CHANGE THIS
set dude_dir = "/mnt/nfs/home/rstein/RotationProject/databases" # should contain decoy.smi and ligand.smi for ROC script 00005...csh
## TO DO - rename this outside in the dir structure and call in blbalbalbabla script
if (-s $dude_dir) then
echo " $dude_dir exist"
else
# this is something to modified in future.
# probably better to exit if it is not there.
echo "databases do not exist. "
echo "consider making a symbolic link to the database files"
#echo "making a symbolic link:"
#echo "ln -s /mnt/nfs/work/users/fischer/VDR/27Jan2014_learningDOCKrgc/databases_all_xtal-ligand_decoy $dude_dir"
#ln -s /mnt/nfs/work/users/fischer/VDR/27Jan2014_learningDOCKrgc/databases_all_xtal-ligand_decoy $dude_dir
endif
# change if you want to use a different or consistent dock version
set dock = ${DOCKBASE}/docking/DOCK/bin/dock64
set list = "2HYY"
#set list = `cat $1`
#set list = `cat file`
# CHANGE THIS (pdbname)
foreach pdbname ( $list )
# creates "ligands" and "decoys" and has the aim to dock all of the subsets for those two
foreach db_type ( "ligands" "decoys" )
set workdir1 = "${mountdir}/${pdbname}/ligands-decoys_sphmod/${db_type}"
mkdir -p ${workdir1}
cd ${workdir1}
# puts dockfiles in the right relative-path that INDOCK file expects
ln -s $filedir/${pdbname}/dockfiles .
set count = '1'
# loop over database files to put each into a seperate chunk
foreach dbfile (`ls $dude_dir/${db_type}/${db_type}*.db2.gz`)
echo $dbfile
set chunk = "chunk$count"
set workdir2 = ${workdir1}/$chunk
## so you don't blow away stuff
if ( -s $workdir2 ) then
echo "$workdir2 exits"
continue
endif
#rm -rf ${workdir}
mkdir -p ${workdir2}
cd ${workdir2}
# copy INDOCK file of choice in right location
#cp $filedir/zzz.dock3_input/INDOCK .
#cp $filedir/INDOCK_match20K INDOCK
#cp $filedir/INDOCK_5k_TolerantClash INDOCK # CHANGE THIS
cp $filedir/${pdbname}/INDOCK .
# modified the dock file using sed. here we change some key sampling parameters; sed -i changes input file internally (overwrites), -e changes file externally (pipes it to screen or into file if redirected)
#sed -i "s/bump_maximum 50.0/bump_maximum 500.0/g" INDOCK
#sed -i "s/bump_rigid 50.0/bump_rigid 500.0/g" INDOCK
#sed -i "s/check_clashes yes/check_clashes no/g" INDOCK
sed -i "s/receptor_sphere_file ..\/dockfiles\/matching_spheres.sph/receptor_sphere_file ..\/..\/..\/working\/matching_spheres_mod.sph/g" INDOCK
ln -s $dbfile .
set dbf = `ls *.gz`
echo "./$dbf"
# says what to dock and where it sits
echo "./$dbf" > split_database_index\
# writes submission script that runs dock on the sgehead queue
cat <<EOF > DOCKING_${db_type}.csh
#\$ -S /bin/csh
#\$ -cwd
#\$ -q all.q
#\$ -o stdout
#\$ -e stderr
cd ${workdir2}
echo "starting . . ."
date
echo $dock
$dock
date
echo "finished . . ."
EOF
qsub DOCKING_${db_type}.csh
# alternatively if you don't want to run it on the queue but locally comment in this instead:
#csh DOCKING_${lig_type}.csh &
@ count = ${count} + 1
# counter is chuch dir
end # dbfile
end # db_type
end # pdbname
make the hing region more polar
see the following page: DOCK_3.7_tart
Virtual Screening
database setup
This part of the tutorial is tailored for shoichet lab use. An outside user of dock might need to deviate from what is described.
Go to zinc and select your compounds of interested:
'http://zinc15.docking.org/tranches/home'
This is the tranches page which allows users to select the region of chemical space of interest.
lets select the fragment preset.
on lets download the index file. This file contains the location of each database on our cluster. outside users will need to download the databases themselves.
now lets setup the directorys for docking by running the following script:
python /nfs/home/tbalius/zzz.github/DOCK/docking/setup/setup_db2_zinc15_file_number.py ./ vs_frag /nfs/work/tbalius/database_ph4/frags.txt 500 count
Note that this script is avable in later beta versons of DOCK3.7.
submitting the docking calculations
This script will submit a job to the queue for each of the docking directorys created by the setup script.
$DOCKBASE/docking/submit/submit.csh
DOCK3.7 is a serial program and is parallelized by submiting many serial jobs to the queue.
combining the results
After your docking jobs have all completed, This script will combine all your results into an extract_all file.
$DOCKBASE/analysis/extract_all.py
This script will create a mol2 file with the top scoring molecules:
$DOCKBASE/analysis/getposes.py
curating and hit-picking
Typically we will visualize the ligands in UCSF Chimera using the veiwdock tool (using the DOCK4, 5, or 6 format).