FlexPepDock: Difference between revisions
Oliv Eidam (talk | contribs) No edit summary |
Oliv Eidam (talk | contribs) No edit summary |
||
| Line 112: | Line 112: | ||
./rescore_batch.csh | ./rescore_batch.csh | ||
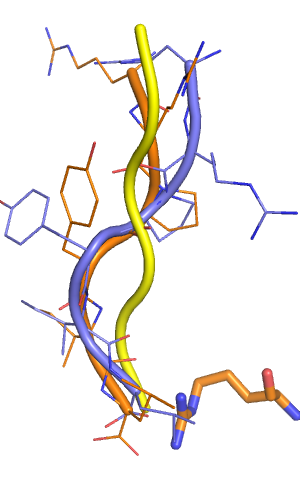
In my prospective prediction, the orange output model had a backbone RMSD of 1.9 Angstroem (3.4 A over HA all atoms), which is a freaking good prediction. The the backbones align perfectly over the last four amino acids, and the two arginines at the N-terminus are actually (and surprisingly) not perfectly ordered in the crystal structure. <br> | |||
As a side note: a few models had RMSD values as low as 1.2 A, but scored worse. | |||
Revision as of 23:46, 7 May 2013
FlexPepDock: Wanna dock a peptide? Use FlexPepDock!
Interested in peptide docking? Use FlexPepDock, which is a peptide docking software implemented in Rosetta. The easiest way to do that is to use the online server:
http://flexpepdock.furmanlab.cs.huji.ac.il
If you want to run FlexPepDock locally, follow the steps below.
In my example I docked the hexapeptide NT(8-13) with the sequence RRPYIL into the neurotensin receptor. I did this prospectively, without having looked at the bound peptide conformation before doing this. All I knew was that the C-terminal carboxylate interacts with Arg327 (Barroso et al, JBC, 2000) and that the peptide adopts an extended conformation (Luca et al, PNAS, 2003).
1. Create peptide coordinates in Pymol
You can generate peptide coordinates using Pymol (Build->Residue->Alanine). If you want an extended peptide, it is best to generate coordinates for poly-Ala first, and then mutate to your peptide using the mutation wizard (Wizard->Mutagenesis->Mutate to Arg->Apply).
Save molecule.
Do NOT add a amine at the N-terminus and do NOT add a C-terminal carboxylate to the pymol coordinates!
Important note: FlexPepDock does not change your Psi angles, so make sure that a proline is in trans if that's what you want!!
2. Dock peptide coordinates in Pymol to generate a starting model for FlexPepDock
You need a rough starting peptide model for FlexPepDock. The peptide starting model should be within 5 Angstroem RMSD to the native structure. You can manually dock your peptide created above in Pymol into to the receptor by switching to Editing Mode, and dragging/rotating the peptide by holding the Shift-Button and Middle and Left-Mouse button, respectively. Don't worry about clashes of the peptide with the protein: the most important thing is that the backbone is within 5 Anstroem of the native structure.
Save the docked peptide coordinates and add them at the below of the PDB coordinates of the apo receptor structure. It is important to add them below the receptor coordinates, separated by a TER statement. Also, there should be no END statements.
3. Run prepack.sh
Run prepack.sh (see code below):
./prepack.sh NTS1_rrpyil_input.pdb
prepack.sh generates pNTS1_rrpyil_input_0001.pdb as output: the protein is protonated and a N-terminal amine and a C-terminal carboxylate have been added to the peptide.
Code of prepack.sh:
#!/bin/csh #$1 is the start.pdb set arch = `uname -p` if ( $arch == 'x86_64') then ~londonir/rosetta/rosetta_source/bin/FlexPepDocking.static.linuxgccrelease -s $1 -database /raid1/people/londonir/rosetta/rosetta_database -native $1 -flexpep_prepack -ex1 -ex2aro -unboundrot $1 > log.prepack else echo "I can only run on a x86_64 system..." endif
4. Run FlexPepDock on Cluster using submit_fpdock.sh
Run FlexPepDock on Cluster using submit_fpdock.sh (see code below):
./submit_fpdock.sh NTS1_rrpyil_input_0001.pdb 200
submit_fpdock.sh takes two arguments: the protonated protein-peptide input model from prepack.sh (NTS1_rrpyil_input_0001.pdb) and the second argument (200) is the number of models you want to generate - 200 is a good number. submit_fpdock.sh calls a script called single_fpdock.sh (see code below), which executes the actual peptide docking. The peptide docking is pretty fast: it takes about 2 minutes per model.
Code of submit_fpdock.sh:
#!/bin/csh #$1 is the start file (start.pdb) #$2 is the number of jobs to send (each will attempt nstruct=1) foreach i (`seq 1 $2`) qsub -l arch=lx24-amd64 -q all.q -cwd -e ./error -o ./out -v arg1=$i,arg2=$1 ./single_fpdock.sh end
Code of single_fpdock.sh:
#!/bin/csh #arg1 is the run prefix number #arg2 is the start.pdb (and native.pdb) set arch = `uname -p` if ( $arch == 'x86_64') then ~londonir/rosetta/rosetta_source/bin/FlexPepDocking.static.linuxgccrelease -s $arg2 -database /raid1/people/londonir/rosetta/rosetta_database -native $arg2 -pep_refine -ex1 -ex2aro -use_input_sc -nstruct 1 -unboundrot $arg2 -out:prefix $arg1'.' > log.$arg1 endif
5. Analyze output models
FlexPepDock outputs a pdb, a score file (.sc) and a log file (log) for each model requested. The score files is a tab-delimited file with many scores and other interesting numbers like Interface buried surface area (I_bsa), number of hydrogen bonds (I_hb), score for the Interface (I_sc) and a score for the peptide (pep_sc). Your main interest is most likely the total score (total_score).
You can print the total scores of each model using this script:
#!/bin/csh
# prints total scores for each model
foreach file (*.sc)
echo $file":" `awk 'NR==3 {print $2}' $file`
end
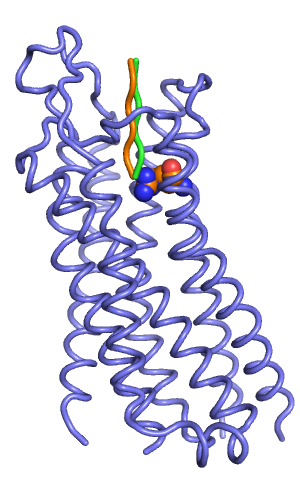
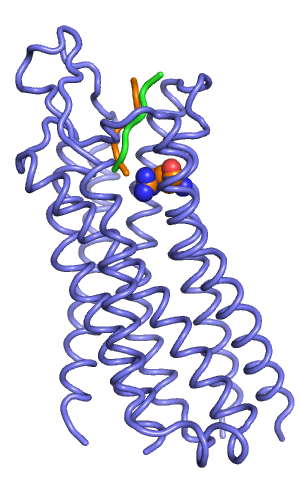
In my prospective peptide docking, the best scoring output model starting from the orange pose had a total score of -324, which was better than the score of the green starting pose (-316).
6. Calculate RMSD values compared to native structure
FlexPepDock automatically outputs RMSD values of the models compared to the input structure (rmsALL, rmsBB etc.). But if you know the native structure, you are probably interested how the models compare to the native structure.
You can calculate RMSD values for each model compared to the native structure using the script rescore_batch.csh. First, you have to generate a list of pdb IDs for which you want to generate RMSDs compared to the native structure:
ls -1 *.pdb > list_of_pdbs.txt
Then run rescore_batch.csh (see code below):
./rescore_batch.csh
In my prospective prediction, the orange output model had a backbone RMSD of 1.9 Angstroem (3.4 A over HA all atoms), which is a freaking good prediction. The the backbones align perfectly over the last four amino acids, and the two arginines at the N-terminus are actually (and surprisingly) not perfectly ordered in the crystal structure.
As a side note: a few models had RMSD values as low as 1.2 A, but scored worse.
Code of rescore_batch.csh:
#!/bin/csh #-s starting #-native: reference for rmsd calc # -l list file ~londonir/rosetta/rosetta_source/bin/FlexPepDocking.static.linuxgccrelease -l list_of_pdbs.txt -database /raid1/people/londonir/rosetta/rosetta_database -native 4GRV_AB_ATOM.pdb -out:prefix rescore_list'.' -flexpep_score_only -out:pdb false > log.rescore_list #create list with pdbs # ls -1 *.pdb > list_of_pdbs.txt
This script takes a few minutes to run and outputs a file called log.rescore_list. For each model, you can print the scores, backbone RMSDs and model name, sorted by backbone RMSD, by typing this command:
awk '{print $2, $41, $53}' rescore_list.score.sc | sort -rgk 2
Acknowlegements
Many thanks to Nir for the introduction into FlexPepDock.