FEP+ for GPCR: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
* Equilibration of complex structure (with confident binding pose) | |||
Carefully look into the binding site and make sure the residues are correctly protonated | |||
Prepare the system: build the POPC membrane, add salts, add solvent | |||
* Force field builder | |||
Run force field builder for all ligands | |||
* Flexible ligand alignment OR core constrain docking | |||
Depends on how similar/different are the ligands to the reference/center ligand | |||
* Create FEP maps | |||
* Write out the submission file; change host; submit on gimel5 via slurm | |||
Revision as of 18:54, 26 February 2021
2/25/2021 Ying Yang
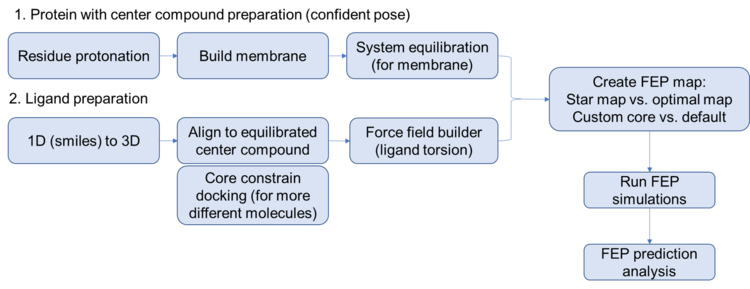
Steps for setting up a FEP prediction for membrane protein
- Equilibration of complex structure (with confident binding pose)
Carefully look into the binding site and make sure the residues are correctly protonated Prepare the system: build the POPC membrane, add salts, add solvent
- Force field builder
Run force field builder for all ligands
- Flexible ligand alignment OR core constrain docking
Depends on how similar/different are the ligands to the reference/center ligand
- Create FEP maps
- Write out the submission file; change host; submit on gimel5 via slurm