Interaction Filtering: Difference between revisions
No edit summary |
No edit summary |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
This is | This is Interaction Filtering version 1.1 (20200601). Please copy the code to your current directory. | ||
$ cp -r /mnt/nfs/home/sgu/code/interfilter . | |||
'''FIRST AND FOREMOST, if you find any version conflicts between python3 and python2, you need to comment out inside ~/.cshrc:''' # source /nfs/soft/dock/versions/dock37/DOCK-3.7-trunk/env.csh | |||
To run the code, you need OpenEye (version 2019.Oct.2) | To run the code, you need to install OpenEye (version 2019.Oct.2) by following the instruction: https://docs.eyesopen.com/toolkits/python/quickstart-python/linuxosx.html | ||
On our cluster, you may source my environment. | On our cluster, you may source my environment. | ||
$ source /nfs/home/sgu/anaconda3/etc/profile.d/conda. | $ source /nfs/home/sgu/anaconda3/etc/profile.d/conda.sh | ||
$ conda activate oepython | $ conda activate oepython | ||
$ | $ source /nfs/soft/openeye/OpenEye-toolkits-python3-linux-x64-2019.10.2/env.sh | ||
Running the code: | Running the code: | ||
$ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2 | $ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2 | ||
If you want to | If you want to have/avoid the interaction (hydrogen bond or salt bridge) for a specific residue (e.g. ASP115A, A means chain A): | ||
In rec.crg.pdb, some residue like HIS is converted to HID or HIE. Please use the converted name instead of HIS. | |||
$ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2 -residue ASP115A | $ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2 -residue ASP115A | ||
If you want to plot the paired/unpaired interaction (figure generation can be slow, but tens of thousands should be fine): | If you want to plot the paired/unpaired interaction (figure generation can be slow, but tens of thousands should be fine): | ||
| Line 27: | Line 29: | ||
To filter out compounds, you may be interested in: | To filter out compounds, you may be interested in: | ||
'''Column3: #hydrogen bond clash + #salt bridge clash | '''Column3: #hydrogen bond clash + #salt bridge clash''' | ||
'''Column4: #unpaired ligand donor + #unpaired ligand salt bridge''' | |||
'''Column5: #unpaired ligand acceptor''' | |||
'''Column9: #hydrogen bond''' | |||
'''Column11: #salt bridge''' | |||
'''Column15: interaction (hydrogen bond or salt bridge) for you specified residue (0 means no, 1 means yes)''' | |||
$ awk '$3==0 && $4==0 && $5<=3 && $9+$11>0' poses_noResidue_interaction_analysis.txt > filtered.txt | |||
$ awk '$3==0 && $4==0 && $5<=3 && $9+$11>0 && $15>0' poses_ASP115A_interaction_analysis.txt > filtered.txt | |||
--------------------------------------------------------------------------------------------------------------------------------------------- | |||
8/5/2020 Ying | |||
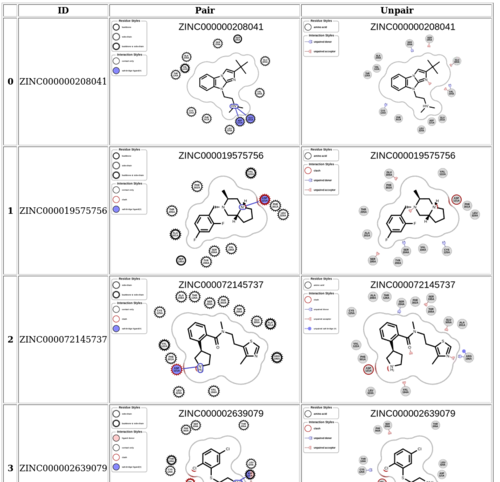
Collect 2D interaction plots into a html file: | |||
cd <folder with png files> | |||
python /nfs/home/yingyangg/scripts/plot_2Dinteraction_html.py | |||
2Dinteraciton.html will be generated, and can be further converted to PDF via print. | |||
[[File: 2d_html.png|thumb|center|500px|2Dinteraction_html plot]] | |||
Latest revision as of 01:54, 18 August 2021
This is Interaction Filtering version 1.1 (20200601). Please copy the code to your current directory.
$ cp -r /mnt/nfs/home/sgu/code/interfilter .
FIRST AND FOREMOST, if you find any version conflicts between python3 and python2, you need to comment out inside ~/.cshrc: # source /nfs/soft/dock/versions/dock37/DOCK-3.7-trunk/env.csh
To run the code, you need to install OpenEye (version 2019.Oct.2) by following the instruction: https://docs.eyesopen.com/toolkits/python/quickstart-python/linuxosx.html
On our cluster, you may source my environment.
$ source /nfs/home/sgu/anaconda3/etc/profile.d/conda.sh $ conda activate oepython $ source /nfs/soft/openeye/OpenEye-toolkits-python3-linux-x64-2019.10.2/env.sh
Running the code:
$ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2
If you want to have/avoid the interaction (hydrogen bond or salt bridge) for a specific residue (e.g. ASP115A, A means chain A):
In rec.crg.pdb, some residue like HIS is converted to HID or HIE. Please use the converted name instead of HIS.
$ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2 -residue ASP115A
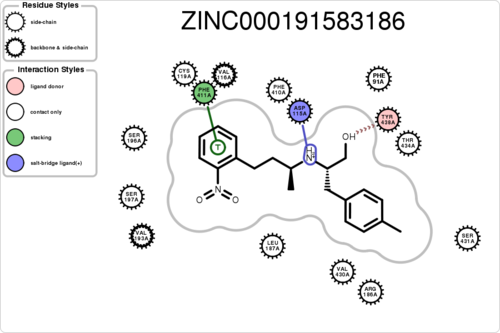
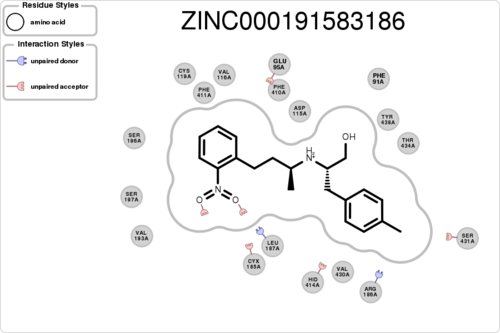
If you want to plot the paired/unpaired interaction (figure generation can be slow, but tens of thousands should be fine):
$ python interfilter.py -protein rec.crg.pdb -ligand poses.mol2 -residue ASP115A -plot
The output is a txt file, containing 15 columns: 1ligand 2clash 3hbond_clash+sbridge_clash 4unpairedl_donor+unpairedl_sbridge 5unpairedl_acceptor 6unpairedp_donor+unpairedp_acceptor 7unpairedp_sbridge 8contact 9hbond+hbond_nonideal 10hbond_ligand 11sbridge 12stacking 13cationpi 14halogen 15residue
To filter out compounds, you may be interested in:
Column3: #hydrogen bond clash + #salt bridge clash
Column4: #unpaired ligand donor + #unpaired ligand salt bridge
Column5: #unpaired ligand acceptor
Column9: #hydrogen bond
Column11: #salt bridge
Column15: interaction (hydrogen bond or salt bridge) for you specified residue (0 means no, 1 means yes)
$ awk '$3==0 && $4==0 && $5<=3 && $9+$11>0' poses_noResidue_interaction_analysis.txt > filtered.txt $ awk '$3==0 && $4==0 && $5<=3 && $9+$11>0 && $15>0' poses_ASP115A_interaction_analysis.txt > filtered.txt
8/5/2020 Ying
Collect 2D interaction plots into a html file:
cd <folder with png files> python /nfs/home/yingyangg/scripts/plot_2Dinteraction_html.py
2Dinteraciton.html will be generated, and can be further converted to PDF via print.