Another getposes.py: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
5/8/2020 Ying | 5/8/2020 Ying | ||
Getting more than one pose | |||
Getting more than one pose... | |||
Example of getting 3 poses for the top scored 6k molecules: | Example of getting 3 poses for the top scored 6k molecules: | ||
/nfs/home/yingyang/programs/miniconda3/envs/teachopencadd/bin/python \ | /nfs/home/yingyang/programs/miniconda3/envs/teachopencadd/bin/python \ | ||
Revision as of 04:47, 9 May 2020
5/8/2020 Ying
Getting more than one pose...
Example of getting 3 poses for the top scored 6k molecules:
/nfs/home/yingyang/programs/miniconda3/envs/teachopencadd/bin/python \ /nfs/home/yingyangg/scripts/get_poses_multi.py -s extract_all.sort.uniq.txt -n 6000 -p 3 -o pose_top6k_x3.mol2
4/20/2020 Ying
Directly call python also works...
/nfs/home/yingyang/programs/miniconda3/envs/teachopencadd/bin/python \ /nfs/home/yingyangg/scripts/get_poses.py -s extract_all.sort.uniq.txt -n 6000 -o pose_top6k.mol2
3/25/2020 Ying
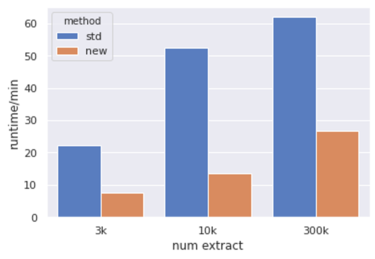
Poses are needed for Shuo's interaction filter and strain filter, sometimes we need to get poses pre-clustering. Owing to the need, here's another get_poses.py script modified on top of getposes_blazing_faster.py from Reed & Trent.
The idea is that we only want to get one pose per zincid with the best dock score. So the script read extract_all.sort.uniq.txt file, and store the min_score for each zincid. When processing mol2.gz file, check if this molecule's mol2 with zincid matches the min_score, otherwise, skip to the next molecule.
First, set environment variable
source /nfs/home/yingyang/.cshrc_opencadd
Get help information:
python /nfs/home/yingyang/scripts/get_poses.py -h
usage: get_poses.py [-h] [-d DIR] [-s SCORE] [-n NUM] [-f FILE] [-o OUT]
[-z GZ_FILE]
optional arguments:
-h, --help show this help message and exit
-d DIR path to where docking is located (default: )
-s SCORE path to where the extract all file is (default:
extract_all.sort.uniq.txt)
-n NUM number of molecules (poses) to get. (default: 500)
-f FILE file contained ligand names to extract (default: None)
-o OUT file name for poses (default: poses.mol2)
-z GZ_FILE file name for input (default: test.mol2.gz)
Example 1, get top 6k molecules from extract_all.sort.uniq.txt (in the docking directory). (getposes routine)
python /nfs/home/yingyangg/scripts/get_poses.py -s extract_all.sort.uniq.txt -n 6000 -o poses_top6k.mol2
Example 2, only get molecules with names listed in a file (for example, zincids of cluster heads), and cut at top 100k.
python /nfs/home/yingyangg/scripts/get_poses.py -s extract_all.sort.uniq.txt -n 100000 -f <zincid.txt> -o poses_interested.mol2
Comparing the computation time: