Screen3d: Difference between revisions
Oliv Eidam (talk | contribs) No edit summary |
Oliv Eidam (talk | contribs) No edit summary |
||
| Line 45: | Line 45: | ||
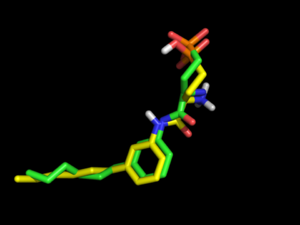
However, changing OMEGA force field to mmff94s with electrostatics [setting omega.SetBuildForceField('mmff94s') and omega.SetSearchForceField('mmff94s') in omega.parm] will produce only 69 conformations, but all of them exhibit the internal salt bridge and one of them has a 3D Tanimoto coefficient of 0.80. | However, changing OMEGA force field to mmff94s with electrostatics [setting omega.SetBuildForceField('mmff94s') and omega.SetSearchForceField('mmff94s') in omega.parm] will produce only 69 conformations, but all of them exhibit the internal salt bridge and one of them has a 3D Tanimoto coefficient of 0.80. | ||
==Acknowledgments== | |||
Thanks to Nir for pointing me to this cool ChemAxon tool. | |||
Revision as of 23:33, 6 May 2013
Sceen3d: Align ligands in 3D
This tool is useful if you want to align (superpose) conformations of a ligand (for example generated by OMEGA). Maybe you are interested to find out if a bioactive conformation of a ligand as represented in a crystal structure is actually generated by OMEGA.
1. Output OMEGA conformations (generated by dbgen.csh) into mol2 file
To output OMEGA conformations generated by dbgen.csh into mol2 file, run the following commands:
# tell dbgen.csh to skip file deletion: setenv DEBUG 1 # run dbgen.csh on your xtal-ligand: dbgen.csh xtal-lig1-.ism # copy the mol2 file with all OMEGA conformations generated for your xtal-ligand: cp ref/TEMP00000001.mol2/3_1.1.save.mol2 # count the number of conformations: grep -c TEMP00000001 3_1.1.save.mol2
2. Superpose OMEGA conformations on xtal-ligand
Use screen3d (ChemAxon) to align the OMEGA conformations with your xtal-ligand:
xtal-lig1-.ism: CCCCCCc1cccc(c1)NC(=O)[C@@H](CCP(=O)([O-])[OH])[NH3+] C38190873
# convert xtal-lig.pdb to mol2 format: convert.py --i=xtal-lig.pdb --o=xtal-lig.mol2 --add_hyd # in this case convert.py creates a double deprotonated phosphonate, # but it does not matter for this analysis, as screen3d will simply # ignore the one phosphonate hydrogen in 3_1.1.save.mol2
# create .ser file for xtal-ligand: screen3d g xtal-lig.mol2 -match # create .ser file for OMEGA conformations: screen3d g 3_1.1.save.mol2 -match # superpose OMEGA conformations on xtal-ligand: screen3d s -t 3_11save.ser -q xtal-lig.ser -match -oformat mol2 # sort output file by 3D Tanimoto: sort -gk 3 3_11save_screenOut.txt
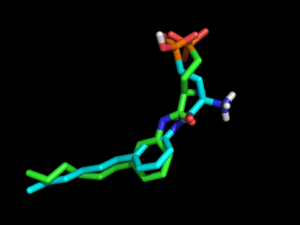
Result: the highest 3D Tanimoto coefficient is 0.64.
In the used example (xtal-lig.pdb is the co-crystallized antagonist of PDB coordinates 3V2Y), no OMEGA conformation exhibits the internal salt bridge between the amine and the phosphonate.
However, changing OMEGA force field to mmff94s with electrostatics [setting omega.SetBuildForceField('mmff94s') and omega.SetSearchForceField('mmff94s') in omega.parm] will produce only 69 conformations, but all of them exhibit the internal salt bridge and one of them has a 3D Tanimoto coefficient of 0.80.
Acknowledgments
Thanks to Nir for pointing me to this cool ChemAxon tool.