Global Matching Sphere Optimization: Difference between revisions
No edit summary |
mNo edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
* a Bash script, that watches created directory structure, runs docking and processes docking results | * a Bash script, that watches created directory structure, runs docking and processes docking results | ||
== Setup & Running == | == Setup & Running == | ||
So far, the program is running on Wynton. LMK if you are interested in launching it on | So far, the program is running on Wynton and Gimel. LMK if you are interested in launching it on other clusters. | ||
The scripts and example config file are in <code>/wynton/group/bks/soft/juggler</code> | The scripts and example config file are in | ||
Wynton | |||
<code>/wynton/group/bks/soft/juggler</code> | |||
Gimel | |||
<code>/nfs/home/ak87/PROGRAM/juggler</code> | |||
=== Setup === | |||
Install [https://wiki.docking.org/index.php?title=SUBDOCK_DOCK3.8 SUBDOCK]<syntaxhighlight lang="bash"> | |||
git clone https://github.com/docking-org/SUBDOCK.git | |||
</syntaxhighlight>Install [https://wiki.docking.org/index.php/Docking_Analysis_in_DOCK3.8#top_poses.py top_poses.py]<syntaxhighlight lang="bash"> | |||
git clone https://github.com/docking-org/docktop.git | |||
</syntaxhighlight> | |||
=== Preparation === | === Preparation === | ||
What you need to prepare: | |||
* <code>dockfiles</code> directory with any tools of your liking (blastermaster, dockopt etc). You will also need | |||
* <code>rec.pdb</code> | |||
* <code>rec.crg.pdb</code> | |||
* <code>xtal-lig.pdb</code>: To get RMSD of xtal-lig docked poses to the experimental pose, your xtal-lig.pdb must have correct bond orders and atom valences. You can edit it in Schrodinger and save as <code>xtal-lig.pdb</code> | |||
* <code>ligands.names</code> | |||
* <code>decoys.names</code> | |||
* <code>sdi</code> file with the paths to ligand .tgz files.. | |||
Prepare <code>juggler_config.yml</code> file. Queue type is <code>sge</code> for Wynton and <code>slurm</code> for Gimel (newer machines, like gimel5/gimel2/n-1-XXX...). Put the config into an empty directory.<syntaxhighlight lang="yaml"> | |||
################################################ | |||
# Paths for your target | |||
# NSP14 -- example | |||
receptor_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NEOCORTEX/SCRIPTS/GENERATION/rec.pdb" | |||
rec_crg_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NEOCORTEX/SCRIPTS/GENERATION/rec.crg.pdb" | |||
xtal_lig_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NEOCORTEX/SCRIPTS/GENERATION/TMP/xtal-lig-no-ring.pdb" | |||
dock_files_dir_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/RECEPTOR/TYR368_0.2-ALA353_0.2-GLY333_0.2/ZINC611-XTAL/LSD/ALL-SPH/dockfiles" | |||
lig_names_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/LIGANDS/REPACK/ligands.names" | |||
dec_names_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/LIGANDS/REPACK/decoys.names" | |||
sdi_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/LIGANDS/REPACK/ligands_repack_wynton_sdi" | |||
################################################ | |||
# Executables and running | |||
dockbase: "/wynton/group/bks/soft/DOCK" | |||
subdock_bash_file_path: "/wynton/home/irwin/ak87/PROGRAM/SUBDOCK/subdock.bash" | |||
queue_type: "sge" # "slurm" or "sge" | |||
top_poses_file: "/wynton/home/irwin/ak87/PROGRAM/docktop/top_poses.py" | |||
############################################### | |||
# Max and min number of spheres | |||
min_sph: 4 # min is 4 | |||
max_sph: 10 # max is 100 | |||
############################################### | |||
# Parameters for genetic algorithm | |||
# Don't need to be adjusted for most purposes | |||
# sample_size: 20 # Please, note that 1/4 of the sample size survives. | |||
# mutation_prob: 0.01 | |||
# crossover_prob: 0.10 | |||
# go_to_next_gen_prob: 0.30 | |||
# gen_new_prob: 0.01 | |||
############################################### | |||
# Parameters for sphere generation | |||
# Don't need to be adjusted for most purposes | |||
# # changed to 1.0 for compatability with xtal-lig spheres | |||
# close_dist: 1.0 | |||
# far_dist: 5.0 | |||
# min_sph: 4 | |||
# max_sph: 10 | |||
# | |||
</syntaxhighlight> | |||
=== Running === | === Running === | ||
==== Launch Juggler: ==== | |||
Enter a screen environment so your run is not interrupted if you disconnect your SSH session. Then do: | Enter a screen environment so your run is not interrupted if you disconnect your SSH session. Then do: | ||
* Wynton | |||
<code>source /wynton/group/bks/soft/python_envs/python3.8.5.sh</code> | <code>source /wynton/group/bks/soft/python_envs/python3.8.5.sh</code> | ||
<code>python /wynton/group/bks/soft/juggler/juggler.py</code> | <code>python /wynton/group/bks/soft/juggler/juggler-v0.8.py 2&>1 > juggler.log</code> | ||
* Gimel (gimel2/gimel5/n-1-XXX ...) | |||
<code>source /nfs/soft/ian/python3.8.5.sh</code> | |||
<code>python /nfs/home/ak87/PROGRAM/juggler/juggler-v0.8.py 2&>1 > juggler.log</code> | |||
You can detach from the screen (Ctrl-A d). | You can detach from the screen (Ctrl-A d). | ||
==== Launch docking daemon😈: ==== | |||
Being in the same directory, open a new screen. Launch a docking daemon: | |||
* Wynton | |||
<code>source /wynton/group/bks/soft/python_envs/python3.8.5.sh</code> | |||
<code>sh /wynton/group/bks/soft/juggler/rundockd-v0.8.sh</code> | |||
* Gimel (gimel2/gimel5/n-1-XXX ...) | |||
<code>/ | <code>source /nfs/soft/ian/python3.8.5.sh</code> | ||
You can run other calculations | <code>sh /nfs/home/ak87/PROGRAM/juggler/rundockd-v0.8.sh</code> | ||
You can run other calculations in the meantime, as Juggler will track the task IDs that it launched. | |||
The run will take few hours to ~2 days depending on the number of actives and decoys and the load of Wynton. | The run will take few hours to ~2 days depending on the number of actives and decoys and the load of Wynton. | ||
=== Processing results === | === Processing results === | ||
At the end of a run you will get a message that convergence was reachedThe script will print the paths to where three best matching sphere sets are: | |||
* best enrichment | * best enrichment | ||
| Line 53: | Line 143: | ||
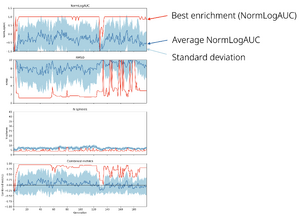
You can also track the optimization progress running the following script in your working directory: | You can also track the optimization progress running the following script in your working directory: | ||
* Wynton | |||
<code>/wynton/group/bks/soft/juggler/plot_all_metrics.py</code> | <code>/wynton/group/bks/soft/juggler/plot_all_metrics.py</code> | ||
* Gimel | |||
<code>/nfs/home/ak87/PROGRAM/juggler/plot_all_metrics.py</code> | |||
It produces <code>combined_metrics.png</code> | It produces <code>combined_metrics.png</code> | ||
| Line 63: | Line 159: | ||
The program will stop after 200 generations if convergence is not reached. In case it takes too long you can stop it any time by pressing Ctrl-C. It doesn't mean that you have no results, though. Juggler generates <code>combined_metrics.dat</code> file in the working directory, which contains metrics for all sets explored. It contains the following columns: | The program will stop after 200 generations if convergence is not reached. In case it takes too long you can stop it any time by pressing Ctrl-C. It doesn't mean that you have no results, though. Juggler generates <code>combined_metrics.dat</code> file in the working directory, which contains metrics for all sets explored. It contains the following columns: | ||
Generations Set# NormLogAUC RMSD Nsph Combined_metrics | <code>Generations Set# NormLogAUC RMSD Nsph Combined_metrics</code> | ||
You can paste its content into Excel, sort by the highest NormLogAUC and pick a MS set of your liking. | You can paste its content into Excel, sort by the highest NormLogAUC and pick a MS set of your liking. | ||
Latest revision as of 00:20, 20 July 2023
Goal
To optimize your matching sphere (MS) setups getting more enrichment with fewer spheres.
Description
The program performs optimization of matching spheres using genetic algorithm. It selects spheres from two sets:
- heavy atoms of xtal-lig
- spheres prepared by SPHGEN program
At each generation, N matching sphere sets are created, containing a maximum of M spheres each. Then retrospective docking is done for each set, and sets are ranked by the
- enrichment (normalized logAUC, see Ian's paper),
- RMSD of the docked pose to the experimental one.
After that, a quarter of sets "survive" and produce a new generation by direct transfer, mutations, and crossover. This process is repeated until enrichment, RMSD and minimum number of spheres do not change substantially in 10 generations.
The program consists of two main modules:
- a Python script (
juggler.py) that performs MS generation, optimization, and ranking. - a Bash script, that watches created directory structure, runs docking and processes docking results
Setup & Running
So far, the program is running on Wynton and Gimel. LMK if you are interested in launching it on other clusters.
The scripts and example config file are in
Wynton
/wynton/group/bks/soft/juggler
Gimel
/nfs/home/ak87/PROGRAM/juggler
Setup
Install SUBDOCK
git clone https://github.com/docking-org/SUBDOCK.git
Install top_poses.py
git clone https://github.com/docking-org/docktop.git
Preparation
What you need to prepare:
dockfilesdirectory with any tools of your liking (blastermaster, dockopt etc). You will also needrec.pdbrec.crg.pdbxtal-lig.pdb: To get RMSD of xtal-lig docked poses to the experimental pose, your xtal-lig.pdb must have correct bond orders and atom valences. You can edit it in Schrodinger and save asxtal-lig.pdbligands.namesdecoys.namessdifile with the paths to ligand .tgz files..
Prepare juggler_config.yml file. Queue type is sge for Wynton and slurm for Gimel (newer machines, like gimel5/gimel2/n-1-XXX...). Put the config into an empty directory.
################################################
# Paths for your target
# NSP14 -- example
receptor_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NEOCORTEX/SCRIPTS/GENERATION/rec.pdb"
rec_crg_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NEOCORTEX/SCRIPTS/GENERATION/rec.crg.pdb"
xtal_lig_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NEOCORTEX/SCRIPTS/GENERATION/TMP/xtal-lig-no-ring.pdb"
dock_files_dir_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/RECEPTOR/TYR368_0.2-ALA353_0.2-GLY333_0.2/ZINC611-XTAL/LSD/ALL-SPH/dockfiles"
lig_names_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/LIGANDS/REPACK/ligands.names"
dec_names_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/LIGANDS/REPACK/decoys.names"
sdi_file_path: "/wynton/home/irwin/ak87/ak87/UCSF/NSP14/LIGANDS/REPACK/ligands_repack_wynton_sdi"
################################################
# Executables and running
dockbase: "/wynton/group/bks/soft/DOCK"
subdock_bash_file_path: "/wynton/home/irwin/ak87/PROGRAM/SUBDOCK/subdock.bash"
queue_type: "sge" # "slurm" or "sge"
top_poses_file: "/wynton/home/irwin/ak87/PROGRAM/docktop/top_poses.py"
###############################################
# Max and min number of spheres
min_sph: 4 # min is 4
max_sph: 10 # max is 100
###############################################
# Parameters for genetic algorithm
# Don't need to be adjusted for most purposes
# sample_size: 20 # Please, note that 1/4 of the sample size survives.
# mutation_prob: 0.01
# crossover_prob: 0.10
# go_to_next_gen_prob: 0.30
# gen_new_prob: 0.01
###############################################
# Parameters for sphere generation
# Don't need to be adjusted for most purposes
# # changed to 1.0 for compatability with xtal-lig spheres
# close_dist: 1.0
# far_dist: 5.0
# min_sph: 4
# max_sph: 10
#
Running
Launch Juggler:
Enter a screen environment so your run is not interrupted if you disconnect your SSH session. Then do:
- Wynton
source /wynton/group/bks/soft/python_envs/python3.8.5.sh
python /wynton/group/bks/soft/juggler/juggler-v0.8.py 2&>1 > juggler.log
- Gimel (gimel2/gimel5/n-1-XXX ...)
source /nfs/soft/ian/python3.8.5.sh
python /nfs/home/ak87/PROGRAM/juggler/juggler-v0.8.py 2&>1 > juggler.log
You can detach from the screen (Ctrl-A d).
Launch docking daemon😈:
Being in the same directory, open a new screen. Launch a docking daemon:
- Wynton
source /wynton/group/bks/soft/python_envs/python3.8.5.sh
sh /wynton/group/bks/soft/juggler/rundockd-v0.8.sh
- Gimel (gimel2/gimel5/n-1-XXX ...)
source /nfs/soft/ian/python3.8.5.sh
sh /nfs/home/ak87/PROGRAM/juggler/rundockd-v0.8.sh
You can run other calculations in the meantime, as Juggler will track the task IDs that it launched.
The run will take few hours to ~2 days depending on the number of actives and decoys and the load of Wynton.
Processing results
At the end of a run you will get a message that convergence was reachedThe script will print the paths to where three best matching sphere sets are:
- best enrichment
- best RMSD
- best balanced metrics (highest enrichment, lowest RMSD and lowest Nsph).
You can use dockfiles from the listed directories.
You can also track the optimization progress running the following script in your working directory:
- Wynton
/wynton/group/bks/soft/juggler/plot_all_metrics.py
- Gimel
/nfs/home/ak87/PROGRAM/juggler/plot_all_metrics.py
It produces combined_metrics.png
If not converged
The program will stop after 200 generations if convergence is not reached. In case it takes too long you can stop it any time by pressing Ctrl-C. It doesn't mean that you have no results, though. Juggler generates combined_metrics.dat file in the working directory, which contains metrics for all sets explored. It contains the following columns:
Generations Set# NormLogAUC RMSD Nsph Combined_metrics
You can paste its content into Excel, sort by the highest NormLogAUC and pick a MS set of your liking.