DOCK 3.7 tutorial based on Webinar 2017/06/28: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
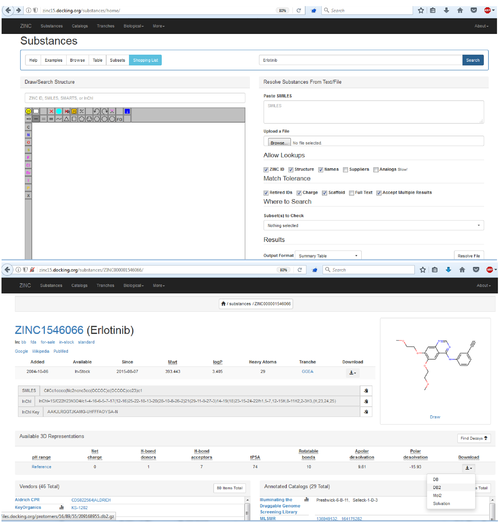
[[File:DOCK3.7tuturial.2017.06.28.png|thumb|center|500px|Search for Your Molecule in ZINC, Get Files for Docking from ZINC]] | [[File:DOCK3.7tuturial.2017.06.28.png|thumb|center|500px|Search for Your Molecule in ZINC, Get Files for Docking from ZINC]] | ||
Get the link from the zinc webpage and us wget to download: | |||
wget http://files.docking.org/protomers/16/89/55/209168955.db2.gz | |||
Put the path of the downloaded database into the split database index file (this file usually contain many db2 file): | |||
ls /path/tutorial_for_webinar/dock3.7/209168955.db2.gz > ligands.sdi | |||
Get the receptor structure from the PDB website | |||
wget https://files.rcsb.org/download/1M17.pdb --no-check-certificate | |||
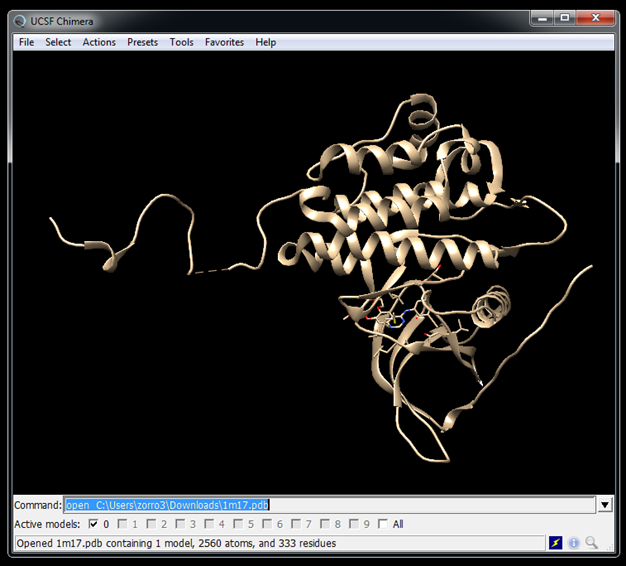
[[File:DOCK3.7tuturial.2017.06.28.2.png]] | |||
Revision as of 20:11, 28 June 2017
Scenario 1: Use docking to predicted how Erlotinib (an approved drug) binds to the Epidermal Growth Factor Receptor
Get the link from the zinc webpage and us wget to download:
wget http://files.docking.org/protomers/16/89/55/209168955.db2.gz
Put the path of the downloaded database into the split database index file (this file usually contain many db2 file):
ls /path/tutorial_for_webinar/dock3.7/209168955.db2.gz > ligands.sdi
Get the receptor structure from the PDB website
wget https://files.rcsb.org/download/1M17.pdb --no-check-certificate